Comparative transcriptional analysis of Persea americana MYB, WRKY and AP2/ERF transcription factors following Phytophthora cinnamomi infection
- PMID: 38590150
- PMCID: PMC11002358
- DOI: 10.1111/mpp.13453
Comparative transcriptional analysis of Persea americana MYB, WRKY and AP2/ERF transcription factors following Phytophthora cinnamomi infection
Abstract
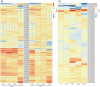
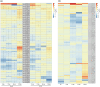
Plant cells undergo extensive transcriptional reprogramming following pathogen infection, with these reprogramming patterns becoming more complex when pathogens, such as hemibiotrophs, exhibit different lifestyles. These transcriptional changes are often orchestrated by MYB, WRKY and AP2/ERF transcription factors (TFs), which modulate both growth and defence-related gene expression. Transcriptional analysis of defence-related genes in avocado (Persea americana) infected with Phytophthora cinnamomi indicated differential immune response activation when comparing a partially resistant and susceptible rootstock. This study identified 226 MYB, 82 WRKY, and 174 AP2/ERF TF-encoding genes in avocado, using a genome-wide approach. Phylogenetic analysis revealed substantial sequence conservation within TF groups underscoring their functional significance. RNA-sequencing analysis in a partially resistant and susceptible avocado rootstock infected with P. cinnamomi was indicative of an immune response switch occurring in either rootstock after 24 and 6 h post-inoculation, respectively. Different clusters of co-expressed TF genes were observed at these times, suggesting the activation of necrotroph-related immune responses at varying intervals between the two rootstocks. This study aids our understanding of avocado immune response activation following P. cinnamomi infection, and the role of the TFs therein, elucidating the transcriptional reprogramming disparities between partially resistant and susceptible rootstocks.
Keywords: Phytophthora; ERF; MYB; WRKY; avocado; defence‐related genes; transcription factor.
© 2024 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures




References
-
- Andam, A. , Azizi, A. , Majdi, M. & Abdolahzadeh, J. (2020) Comparative expression profile of some putative resistance genes of chickpea genotypes in response to ascomycete fungus, Ascochyta rabiei (Pass.) Labr. Brazilian Journal of Botany, 43, 123–130.
-
- Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics: Babraham Institute. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Accessed 11th February 2021]
-
- Backer, R. , Engelbrecht, J. & van den Berg, N. (2022) Differing responses to Phytophthora cinnamomi infection in susceptible and partially resistant Persea americana (Mill.) rootstocks: a case for the role of receptor‐like kinases and apoplastic proteases. Frontiers in Plant Science, 13, 928176. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

