Peeling back the many layers of competitive exclusion
- PMID: 38591029
- PMCID: PMC11000858
- DOI: 10.3389/fmicb.2024.1342887
Peeling back the many layers of competitive exclusion
Abstract
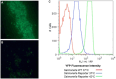
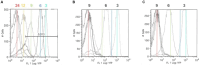
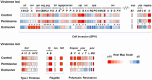
Baby chicks administered a fecal transplant from adult chickens are resistant to Salmonella colonization by competitive exclusion. A two-pronged approach was used to investigate the mechanism of this process. First, Salmonella response to an exclusive (Salmonella competitive exclusion product, Aviguard®) or permissive microbial community (chicken cecal contents from colonized birds containing 7.85 Log10Salmonella genomes/gram) was assessed ex vivo using a S. typhimurium reporter strain with fluorescent YFP and CFP gene fusions to rrn and hilA operon, respectively. Second, cecal transcriptome analysis was used to assess the cecal communities' response to Salmonella in chickens with low (≤5.85 Log10 genomes/g) or high (≥6.00 Log10 genomes/g) Salmonella colonization. The ex vivo experiment revealed a reduction in Salmonella growth and hilA expression following co-culture with the exclusive community. The exclusive community also repressed Salmonella's SPI-1 virulence genes and LPS modification, while the anti-virulence/inflammatory gene avrA was upregulated. Salmonella transcriptome analysis revealed significant metabolic disparities in Salmonella grown with the two different communities. Propanediol utilization and vitamin B12 synthesis were central to Salmonella metabolism co-cultured with either community, and mutations in propanediol and vitamin B12 metabolism altered Salmonella growth in the exclusive community. There were significant differences in the cecal community's stress response to Salmonella colonization. Cecal community transcripts indicated that antimicrobials were central to the type of stress response detected in the low Salmonella abundance community, suggesting antagonism involved in Salmonella exclusion. This study indicates complex community interactions that modulate Salmonella metabolism and pathogenic behavior and reduce growth through antagonism may be key to exclusion.
Keywords: Salmonella; antimicrobials; attenuation; competition; exclusion; pathogen.
Copyright © 2024 Maurer, Cheng, Pedroso, Thompson, Akter, Kwan, Morota, Kinstler, Porwollik, McClelland, Escalante-Semerena and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures






Similar articles
-
Salmonella exploits host- and bacterial-derived β-alanine for replication inside host macrophages.Elife. 2025 Jun 19;13:RP103714. doi: 10.7554/eLife.103714. Elife. 2025. PMID: 40536105 Free PMC article.
-
Eliciting adverse effects data from participants in clinical trials.Cochrane Database Syst Rev. 2018 Jan 16;1(1):MR000039. doi: 10.1002/14651858.MR000039.pub2. Cochrane Database Syst Rev. 2018. PMID: 29372930 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
-
The effectiveness of selected feed and water additives for reducing Salmonella spp. of public health importance in broiler chickens: a systematic review, meta-analysis, and meta-regression approach.Prev Vet Med. 2012 Oct 1;106(3-4):197-213. doi: 10.1016/j.prevetmed.2012.07.007. Epub 2012 Aug 2. Prev Vet Med. 2012. PMID: 22858425
-
Distinct adaptation and epidemiological success of different genotypes within Salmonella enterica serovar Dublin.Elife. 2025 Jun 25;13:RP102253. doi: 10.7554/eLife.102253. Elife. 2025. PMID: 40560760 Free PMC article.
Cited by
-
Longitudinal study highlights patterns of Salmonella serovar co-occurrence and exclusion in commercial poultry production.Front Microbiol. 2025 Jul 16;16:1570593. doi: 10.3389/fmicb.2025.1570593. eCollection 2025. Front Microbiol. 2025. PMID: 40740319 Free PMC article.
References
-
- Anonymous (2021). Salmonella outbreak linked to wild songbirds. Center for Disease Control and Prevention. Available at: https://www.cdc.gov/salmonella/typhimurium-04-21/index.html (Accessed April 29, 2021).
-
- Azcarate-Peril M. A., Butz N., Cadenas M. B., Koci M., Ballou A., Mendoza M., et al. . (2018). An attenuated Salmonella enterica Serovar typhimurium strain and Galacto-oligosaccharides accelerate clearance of Salmonella infections in poultry through modifications to the gut microbiome. Appl. Environ. Microbiol. 84:e02526-17. doi: 10.1128/AEM.02526-17, PMID: - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

