Genetic Testing for Supravalvar Aortic Stenosis: What to Do When It Is Not Williams Syndrome
- PMID: 38591341
- PMCID: PMC11262489
- DOI: 10.1161/JAHA.123.034048
Genetic Testing for Supravalvar Aortic Stenosis: What to Do When It Is Not Williams Syndrome
Abstract
Background: We aimed to describe the frequency and yield of genetic testing in supravalvar aortic stenosis (SVAS) following negative evaluation for Williams-Beuren syndrome (WS).
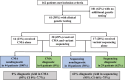
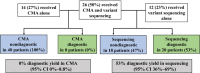
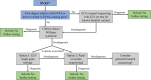
Methods and results: This retrospective cohort study included patients with SVAS at our institution who had a negative evaluation for WS from May 1991 to September 2021. SVAS was defined as (1) peak supravalvar velocity of ≥2 meters/second, (2) sinotubular junction or ascending aortic Z score <-2.0, or (3) sinotubular junction Z score <-1.5 with family history of SVAS. Patients with complex congenital heart disease, aortic valve disease as the primary condition, or only postoperative SVAS were excluded. Genetic testing and diagnoses were reported. Of 162 patients who were WS negative meeting inclusion criteria, 61 had genetic testing results available (38%). Chromosomal microarray had been performed in 44 of 61 and was nondiagnostic for non-WS causes of SVAS. Sequencing of 1 or more genes was performed in 47 of 61. Of these, 39 of 47 underwent ELN sequencing, 20 of 39 (51%) of whom had a diagnostic variant. Other diagnoses made by gene sequencing were Noonan syndrome (3 PTPN11, 1 RIT1), Alagille syndrome (3 JAG1), neurofibromatosis (1 NF1), and homozygous familial hypercholesterolemia (1 LDLR1). Overall, sequencing was diagnostic in 29 of 47 (62%).
Conclusions: When WS is excluded, gene sequencing for SVAS is high yield, with the highest yield for the ELN gene. Therefore, we recommend gene sequencing using a multigene panel or exome analysis. Hypercholesterolemia can also be considered in individuals bearing the stigmata of this disease.
Keywords: SVAS; Williams syndrome; cardiovascular; genetics.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

