ZNF397 Deficiency Triggers TET2-Driven Lineage Plasticity and AR-Targeted Therapy Resistance in Prostate Cancer
- PMID: 38591846
- PMCID: PMC11285331
- DOI: 10.1158/2159-8290.CD-23-0539
ZNF397 Deficiency Triggers TET2-Driven Lineage Plasticity and AR-Targeted Therapy Resistance in Prostate Cancer
Abstract
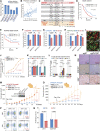
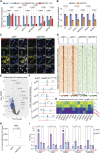
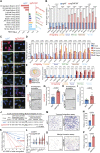
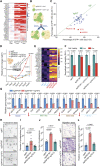
Cancer cells exhibit phenotypical plasticity and epigenetic reprogramming that allows them to evade lineage-dependent targeted treatments by adopting lineage plasticity. The underlying mechanisms by which cancer cells exploit the epigenetic regulatory machinery to acquire lineage plasticity and therapy resistance remain poorly understood. We identified zinc finger protein 397 (ZNF397) as a bona fide coactivator of the androgen receptor (AR), essential for the transcriptional program governing AR-driven luminal lineage. ZNF397 deficiency facilitates the transition of cancer cell from an AR-driven luminal lineage to a ten-eleven translocation 2 (TET2)-driven lineage plastic state, ultimately promoting resistance to therapies inhibiting AR signaling. Intriguingly, our findings indicate that a TET2 inhibitor can eliminate the resistance to AR-targeted therapies in ZNF397-deficient tumors. These insights uncover a novel mechanism through which prostate cancer acquires lineage plasticity via epigenetic rewiring and offer promising implications for clinical interventions designed to overcome therapy resistance dictated by lineage plasticity. Significance: This study reveals a bifurcated role of ZNF397, and a TET2-driven epigenetic mechanism regulating tumor lineage plasticity and therapy response in prostate cancer, enhances the understanding of drug resistance, and unveils a new therapeutic strategy for overcoming androgen receptor-targeted therapy resistance.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
M. Sjöström reports grants from the Prostate Cancer Foundation during the conduct of the study. Y. Chen reports grants from NCI and Prostate Cancer Foundation during the conduct of the study; grants from Foghorn Pharmacetuicals and personal fees and other support from ORIC Pharmaceuticals outside the submitted work. A.B. Hanker reports grants from NIH/NCI during the conduct of the study; grants from Breast Cancer Research Foundation, non-financial support from Tempus and Daiichi Sankyo; grants from Takeda and Lilly; non-financial support from Puma Biotechnology outside the submitted work. G.V. Raj reports other support from Etirarx, Bayer, and Pfizer; other support from Astellas outside the submitted work. Zhao Wang reports grants from NIGMS R01GM143380, and NHLBI R01HL162842; grants from Welch Fundation Q-2173-20230405 outside the submitted work. C.L. Arteaga reports personal fees from Sanofi, OrigiMed, TAIHO Oncology, AstraZeneca, Daiichi Sankyo, Laekna Therapeutics; grants from Pfizer; personal fees from Komen Foundation outside the submitted work. H. Liang reports H. Liang is a shareholder and scientific advisor of Precision Scientific Ltd. F.Y. Feng reports F.Y. Feng is an advisor and holds equity in Artera, and has served as an advisor to Astellas, Bayer, Blue Earth Diagnostics, BMS, ClearNote, Janssen, Myovant, Point Biopharma, Novartis, Roivant, Sanofi, SerImmune and Amgen. T. Wang reports other support from NightStar Biotechnologies, Inc. and personal fees from Merck, Inc. outside the submitted work. P. Mu reports grants from National Cancer Institute, Department of Defense, Cancer Prevention Research Institute, Prostate Cancer Foundation, and Welch Foundation during the conduct of the study; personal fees from Accutar Biotechnology outside the submitted work. No disclosures were reported by the other authors.
Figures


Update of
-
ZNF397 Loss Triggers TET2-driven Epigenetic Rewiring, Lineage Plasticity, and AR-targeted Therapy Resistance in AR-dependent Cancers.bioRxiv [Preprint]. 2023 Oct 27:2023.10.24.563645. doi: 10.1101/2023.10.24.563645. bioRxiv. 2023. Update in: Cancer Discov. 2024 Aug 2;14(8):1496-1521. doi: 10.1158/2159-8290.CD-23-0539. PMID: 37961351 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- RR170050/Cancer Prevention and Research Institute of Texas (CPRIT)
- R00CA218885/National Cancer Institute (NCI)
- W81XWH-18-1-0411/U.S. Department of Defense (DOD)
- I-2005-20190330/Welch Foundation (The Welch Foundation)
- F31CA261019-01A1/National Cancer Institute (NCI)
- R01 CA292949/CA/NCI NIH HHS/United States
- DDBrown awardee/Life Sciences Research Foundation (LSRF)
- PPG19-1090/Terry Fox Foundation (La Fondation Terry Fox)
- R00 CA218885/CA/NCI NIH HHS/United States
- R01 CA288820/CA/NCI NIH HHS/United States
- R37 CA258730/CA/NCI NIH HHS/United States
- R01 GM143380/GM/NIGMS NIH HHS/United States
- RP230363/Cancer Prevention and Research Institute of Texas (CPRIT)
- 17YOUN12/Prostate Cancer Foundation (PCF)
- 21YOUN10/Prostate Cancer Foundation (PCF)
- P30 CA008748/CA/NCI NIH HHS/United States
- W81XWH21-1-0418/U.S. Department of Defense (DOD)
- F31 CA261019/CA/NCI NIH HHS/United States
- R01CA258584/National Cancer Institute (NCI)
- R01 CA258584/CA/NCI NIH HHS/United States
- T32 CA124334/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

