Siderophore-mediated iron acquisition by Klebsiella pneumoniae
- PMID: 38591913
- PMCID: PMC11112993
- DOI: 10.1128/jb.00024-24
Siderophore-mediated iron acquisition by Klebsiella pneumoniae
Abstract
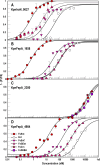
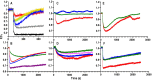
Microbes synthesize and secrete siderophores, that bind and solubilize precipitated or otherwise unavailable iron in their microenvironments. Gram (-) bacterial TonB-dependent outer membrane receptors capture the resulting ferric siderophores to begin the uptake process. From their similarity to fepA, the structural gene for the Escherichia coli ferric enterobactin (FeEnt) receptor, we identified four homologous genes in the human and animal ESKAPE pathogen Klebsiella pneumoniae (strain Kp52.145). One locus encodes IroN (locus 0027 on plasmid pII), and three other loci encode other FepA orthologs/paralogs (chromosomal loci 1658, 2380, and 4984). Based on the crystal structure of E. coli FepA (1FEP), we modeled the tertiary structures of the K. pneumoniae FepA homologs and genetically engineered individual Cys substitutions in their predicted surface loops. We subjected bacteria expressing the Cys mutant proteins to modification with extrinsic fluorescein maleimide (FM) and used the resulting fluorescently labeled cells to spectroscopically monitor the binding and transport of catecholate ferric siderophores by the four different receptors. The FM-modified FepA homologs were nanosensors that defined the ferric catecholate uptake pathways in pathogenic strains of K. pneumoniae. In Kp52.145, loci 1658 and 4984 encoded receptors that primarily recognized and transported FeEnt; locus 0027 produced a receptor that principally bound and transported FeEnt and glucosylated FeEnt (FeGEnt); locus 2380 encoded a protein that bound ferric catecholate compounds but did not detectably transport them. The sensors also characterized the uptake of iron complexes, including FeGEnt, by the hypervirulent, hypermucoviscous K. pneumoniae strain hvKp1.
Importance: Both commensal and pathogenic bacteria produce small organic chelators, called siderophores, that avidly bind iron and increase its bioavailability. Klebsiella pneumoniae variably produces four siderophores that antagonize host iron sequestration: enterobactin, glucosylated enterobactin (also termed salmochelin), aerobactin, and yersiniabactin, which promote colonization of different host tissues. Abundant evidence links bacterial iron acquisition to virulence and infectious diseases. The data we report explain the recognition and transport of ferric catecholates and other siderophores, which are crucial to iron acquisition by K. pneumoniae.
Keywords: ESKAPE pathogen; Klebsiella pneumoniae; TonB-dependent iron acquisition; ferric enterobactin; fluorescent sensors; iron transport; siderophore antibiotic conjugates; siderophores; site-directed mutagenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

