Identification and Characterization of the AREB/ABF Gene Family in Three Orchid Species and Functional Analysis of DcaABI5 in Arabidopsis
- PMID: 38592811
- PMCID: PMC10974128
- DOI: 10.3390/plants13060774
Identification and Characterization of the AREB/ABF Gene Family in Three Orchid Species and Functional Analysis of DcaABI5 in Arabidopsis
Abstract
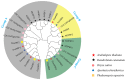
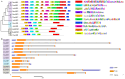
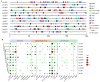
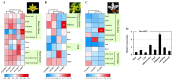
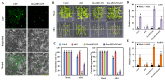
AREB/ABF (ABA response element binding) proteins in plants are essential for stress responses, while our understanding of AREB/ABFs from orchid species, important traditional medicinal and ornamental plants, is limited. Here, twelve AREB/ABF genes were identified within three orchids' complete genomes and classified into three groups through phylogenetic analysis, which was further supported with a combined analysis of their conserved motifs and gene structures. The cis-element analysis revealed that hormone response elements as well as light and stress response elements were widely rich in the AREB/ABFs. A prediction analysis of the orchid ABRE/ABF-mediated regulatory network was further constructed through cis-regulatory element (CRE) analysis of their promoter regions. And it revealed that several dominant transcriptional factor (TF) gene families were abundant as potential regulators of these orchid AREB/ABFs. Expression profile analysis using public transcriptomic data suggested that most AREB/ABF genes have distinct tissue-specific expression patterns in orchid plants. Additionally, DcaABI5 as a homolog of ABA INSENSITIVE 5 (ABI5) from Arabidopsis was selected for further analysis. The results showed that transgenic Arabidopsis overexpressing DcaABI5 could rescue the ABA-insensitive phenotype in the mutant abi5. Collectively, these findings will provide valuable information on AREB/ABF genes in orchids.
Keywords: ABA INSENSITIVE 5 (ABI5); AREB/ABF genes; abscisic acid (ABA); cis-regulatory element (CRE) analysis; gene expression; gene family; orchid.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Identification and expression of the AREB/ABF/ABI5 subfamily genes in chickpea and lentil reveal major players involved in ABA-mediated defense response to drought stress.Planta. 2025 Jun 10;262(1):22. doi: 10.1007/s00425-025-04740-y. Planta. 2025. PMID: 40493071
-
Genome-Wide Identification and Characterization of the AREB/ABF/ABI5 Subfamily Members from Solanum tuberosum.Int J Mol Sci. 2019 Jan 14;20(2):311. doi: 10.3390/ijms20020311. Int J Mol Sci. 2019. PMID: 30646545 Free PMC article.
-
Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants.Physiol Plant. 2013 Jan;147(1):15-27. doi: 10.1111/j.1399-3054.2012.01635.x. Epub 2012 May 16. Physiol Plant. 2013. PMID: 22519646 Review.
-
Identification of ABF/AREB gene family in tomato (Solanum lycopersicum L.) and functional analysis of ABF/AREB in response to ABA and abiotic stresses.PeerJ. 2023 May 4;11:e15310. doi: 10.7717/peerj.15310. eCollection 2023. PeerJ. 2023. PMID: 37163152 Free PMC article.
-
Updates on the Role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA Signaling in Different Developmental Stages in Plants.Cells. 2021 Aug 5;10(8):1996. doi: 10.3390/cells10081996. Cells. 2021. PMID: 34440762 Free PMC article. Review.
Cited by
-
Genomic signatures of SnRKs highlighted conserved evolution within orchids and stress responses through ABA signaling in the Cymbidium ensifolium.BMC Plant Biol. 2025 Mar 3;25(1):277. doi: 10.1186/s12870-025-06280-9. BMC Plant Biol. 2025. PMID: 40025443 Free PMC article.
-
Identification and expression of the AREB/ABF/ABI5 subfamily genes in chickpea and lentil reveal major players involved in ABA-mediated defense response to drought stress.Planta. 2025 Jun 10;262(1):22. doi: 10.1007/s00425-025-04740-y. Planta. 2025. PMID: 40493071
References
-
- Chase M.W., Cameron K.M., Freudenstein J.V., Pridgeon A.M., Salazar G., van den Berg C., Schuiteman A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015;177:151–174. doi: 10.1111/boj.12234. - DOI
-
- Kindlmann P., Kull T., McCormick M. The Distribution and Diversity of Orchids. Diversity. 2023;15:810. doi: 10.3390/d15070810. - DOI
-
- Mérillon J.M., Kodja H. Orchids Phytochemistry, Biology and Horticulture. Springer; Berlin/Heidelberg, Germany: 2019.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

