Genetic Diversity and Population Structure of Maize (Zea mays L.) Inbred Lines in Association with Phenotypic and Grain Qualitative Traits Using SSR Genotyping
- PMID: 38592835
- PMCID: PMC10975177
- DOI: 10.3390/plants13060823
Genetic Diversity and Population Structure of Maize (Zea mays L.) Inbred Lines in Association with Phenotypic and Grain Qualitative Traits Using SSR Genotyping
Abstract
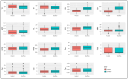
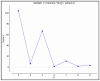
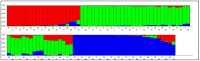
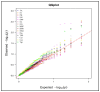
Maize (Zea mays L.) is an important cereal and is affected by climate change. Therefore, the production of climate-smart maize is urgently needed by preserving diverse genetic backgrounds through the exploration of their genetic diversity. To achieve this, 96 maize inbred lines were used to screen for phenotypic yield-associated traits and grain quality parameters. These traits were studied across two different environments (Anand and Godhra) and polymorphic simple sequence repeat (SSR) markers were employed to investigate the genetic diversity, population structure, and trait-linked association. Genotype-environment interaction (GEI) reveals that most of the phenotypic traits were governed by the genotype itself across the environments, except for plant and ear height, which largely interact with the environment. The genotypic correlation was found to be positive and significant among protein, lysine and tryptophan content. Similarly, yield-attributing traits like ear girth, kernel rows ear-1, kernels row-1 and number of kernels ear-1 were strongly correlated to each other. Pair-wise genetic distance ranged from 0.0983 (1820194/T1 and 1820192/4-20) to 0.7377 (IGI-1101 and 1820168/T1). The SSRs can discriminate the maize population into three distinct groups and shortlisted two genotypes (IGI-1101 and 1820168/T1) as highly diverse lines. Out of the studied 136 SSRs, 61 were polymorphic to amplify a total of 131 alleles (2-3 per loci) with 0.46 average gene diversity. The Polymorphism Information Content (PIC) ranged from 0.24 (umc1578) to 0.58 (umc2252). Similarly, population structure analysis revealed three distinct groups with 19.79% admixture among the genotypes. Genome-wide scanning through a mixed linear model identifies the stable association of the markers umc2038, umc2050 and umc2296 with protein, umc2296 and umc2252 with tryptophan, and umc1535 and umc1303 with total soluble sugar. The obtained maize lines and SSRs can be utilized in future maize breeding programs in relation to other trait characterizations, developments, and subsequent molecular breeding performances for trait introgression into elite genotypes.
Keywords: GWAS; SSR; Zea mays; genetic diversity; population structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Slafer G.A., Otegui M.E. Physiological Bases for Maize Improvement. CRC Press; Boca Raton, FL, USA: 2000.
-
- Roychowdhury R., Taoutaou A., Hakeem K.R., Gawwad M.R.A., Tah J. Crop Improvement in the Era of Climate Change. IK International Publishing House; New Delhi, India: 2014. Molecular Marker-Assisted Technologies for Crop Improvement; pp. 241–258.
-
- Prasanna B.M., Cairns J.E., Zaidi P.H., Beyene Y., Makumbi D., Gowda M., Magorokosho C., Zaman-Allah M., Olsen M., Das A. Beat the Stress: Breeding for Climate Resilience in Maize for the Tropical Rainfed Environments. Theor. Appl. Genet. 2021;134:1729–1752. doi: 10.1007/s00122-021-03773-7. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

