Blocking lipid synthesis induces DNA damage in prostate cancer and increases cell death caused by PARP inhibition
- PMID: 38593154
- PMCID: PMC11161871
- DOI: 10.1126/scisignal.adh1922
Blocking lipid synthesis induces DNA damage in prostate cancer and increases cell death caused by PARP inhibition
Abstract
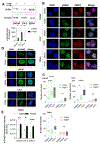
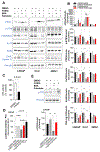
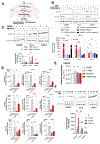
Androgen deprivation therapy (ADT) is the primary treatment for prostate cancer; however, resistance to ADT invariably develops, leading to castration-resistant prostate cancer (CRPC). Prostate cancer progression is marked by increased de novo synthesis of fatty acids due to overexpression of fatty acid synthase (FASN), making this enzyme a therapeutic target for prostate cancer. Inhibition of FASN results in increased intracellular amounts of ceramides and sphingomyelin, leading to DNA damage through the formation of DNA double-strand breaks and cell death. We found that combining a FASNi with the poly-ADP ribose polymerase (PARP) inhibitor olaparib, which induces cell death by blocking DNA damage repair, resulted in a more pronounced reduction in cell growth than that caused by either drug alone. Human CRPC organoids treated with a combination of PARP and FASNi were smaller, had decreased cell proliferation, and showed increased apoptosis and necrosis. Together, these data indicate that targeting FASN increases the therapeutic efficacy of PARP inhibitors by impairing DNA damage repair, suggesting that combination therapies should be explored for CRPC.
Conflict of interest statement
Competing interests
No conflict of interest to be reported.
Figures




References
-
- Migita T, Ruiz S, Fornari A, Fiorentino M, Priolo C, Zadra G, Inazuka F, Grisanzio C, Palescandolo E, Shin E, Fiore C, Xie W, Kung AL, Febbo PG, Subramanian A, Mucci L, Ma J, Signoretti S, Stampfer M, Hahn WC, Finn S, Loda M, Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst 101, 519–532 (2009); published online EpubApr (10.1093/jnci/djp030). - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

