Pharmacological induction of chromatin remodeling drives chemosensitization in triple-negative breast cancer
- PMID: 38593809
- PMCID: PMC11031425
- DOI: 10.1016/j.xcrm.2024.101504
Pharmacological induction of chromatin remodeling drives chemosensitization in triple-negative breast cancer
Abstract
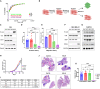
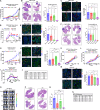
Targeted therapies have improved outcomes for certain cancer subtypes, but cytotoxic chemotherapy remains a mainstay for triple-negative breast cancer (TNBC). The epithelial-to-mesenchymal transition (EMT) is a developmental program co-opted by cancer cells that promotes metastasis and chemoresistance. There are no therapeutic strategies specifically targeting mesenchymal-like cancer cells. We report that the US Food and Drug Administration (FDA)-approved chemotherapeutic eribulin induces ZEB1-SWI/SNF-directed chromatin remodeling to reverse EMT that curtails the metastatic propensity of TNBC preclinical models. Eribulin induces mesenchymal-to-epithelial transition (MET) in primary TNBC in patients, but conventional chemotherapy does not. In the treatment-naive setting, but not after acquired resistance to other agents, eribulin sensitizes TNBC cells to subsequent treatment with other chemotherapeutics. These findings provide an epigenetic mechanism of action of eribulin, supporting its use early in the disease process for MET induction to prevent metastatic progression and chemoresistance. These findings warrant prospective clinical evaluation of the chemosensitizing effects of eribulin in the treatment-naive setting.
Keywords: EMT; breast cancer; chemotherapy; drug resistance; epigenetic; epithelial-to-mesenchymal transition; mesenchymal-to-epithelial transition; metastasis.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.A.M.S. is an employee of Lifebytes India Pvt. Ltd. D.R.P. received research funding from Eisai Pharmaceutical Ltd.
Figures






References
-
- Cortes J., O'Shaughnessy J., Loesch D., Blum J.L., Vahdat L.T., Petrakova K., Chollet P., Manikas A., Diéras V., Delozier T., et al. Eribulin monotherapy versus treatment of physician's choice in patients with metastatic breast cancer (EMBRACE): a phase 3 open-label randomised study. Lancet. 2011;377:914–923. doi: 10.1016/S0140-6736(11)60070-6. - DOI - PubMed
-
- Yoshida T., Ozawa Y., Kimura T., Sato Y., Kuznetsov G., Xu S., Uesugi M., Agoulnik S., Taylor N., Funahashi Y., Matsui J. Eribulin mesilate suppresses experimental metastasis of breast cancer cells by reversing phenotype from epithelial-mesenchymal transition (EMT) to mesenchymal-epithelial transition (MET) states. Br. J. Cancer. 2014;110:1497–1505. doi: 10.1038/bjc.2014.80. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

