Prey killing without invasion by Bdellovibrio bacteriovorus defective for a MIDAS-family adhesin
- PMID: 38594280
- PMCID: PMC11003981
- DOI: 10.1038/s41467-024-47412-3
Prey killing without invasion by Bdellovibrio bacteriovorus defective for a MIDAS-family adhesin
Abstract
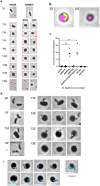
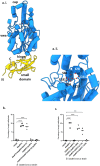
The bacterium Bdellovibrio bacteriovorus is a predator of other Gram-negative bacteria. The predator invades the prey's periplasm and modifies the prey's cell wall, forming a rounded killed prey, or bdelloplast, containing a live B. bacteriovorus. Redundancy in adhesive processes makes invasive mutants rare. Here, we identify a MIDAS adhesin family protein, Bd0875, that is expressed at the predator-prey invasive junction and is important for successful invasion of prey. A mutant strain lacking bd0875 is still able to form round, dead bdelloplasts; however, 10% of the bdelloplasts do not contain B. bacteriovorus, indicative of an invasion defect. Bd0875 activity requires the conserved MIDAS motif, which is linked to catch-and-release activity of MIDAS proteins in other organisms. A proteomic analysis shows that the uninvaded bdelloplasts contain B. bacteriovorus proteins, which are likely secreted into the prey by the Δbd0875 predator during an abortive invasion period. Thus, secretion of proteins into the prey seems to be sufficient for prey killing, even in the absence of a live predator inside the prey periplasm.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

