Bioorthogonal masked acylating agents for proximity-dependent RNA labelling
- PMID: 38594368
- PMCID: PMC11613155
- DOI: 10.1038/s41557-024-01493-1
Bioorthogonal masked acylating agents for proximity-dependent RNA labelling
Abstract
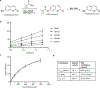
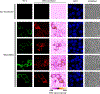
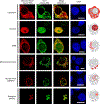
RNA localization is highly regulated, with subcellular organization driving context-dependent cell physiology. Although proximity-based labelling technologies that use highly reactive radicals or carbenes provide a powerful method for unbiased mapping of protein organization within a cell, methods for unbiased RNA mapping are scarce and comparably less robust. Here we develop α-alkoxy thioenol and chloroenol esters that function as potent acylating agents upon controlled ester unmasking. We pair these probes with subcellular-localized expression of a bioorthogonal esterase to establish a platform for spatial analysis of RNA: bioorthogonal acylating agents for proximity labelling and sequencing (BAP-seq). We demonstrate that, by selectively unmasking the enol probe in a locale of interest, we can map RNA distribution in membrane-bound and membrane-less organelles. The controlled-release acylating agent chemistry and corresponding BAP-seq method expand the scope of proximity labelling technologies and provide a powerful approach to interrogate the cellular organization of RNAs.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
B.C.D. is a founder and holds equity in Tornado Bio, Inc. The other authors declare no competing interests.
Figures







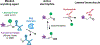



References
-
- Keskin O, Tuncbag N & Gursoy A Predicting protein–protein interactions from the molecular to the proteome level. Chem. Rev. 116, 4884–4909 (2016). - PubMed
-
- Hentze MW, Castello A, Schwarzl T & Preiss T A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018). - PubMed
-
- Engreitz JM, Ollikainen N & Guttman M Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MH122142/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- RM1 HG008935/HG/NHGRI NIH HHS/United States
- F30 DK125088/DK/NIDDK NIH HHS/United States
- GM119840/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- DK125088/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

