IFI35 regulates non-canonical NF-κB signaling to maintain glioblastoma stem cells and recruit tumor-associated macrophages
- PMID: 38594444
- PMCID: PMC11165006
- DOI: 10.1038/s41418-024-01292-8
IFI35 regulates non-canonical NF-κB signaling to maintain glioblastoma stem cells and recruit tumor-associated macrophages
Abstract
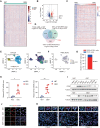
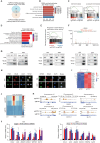
Glioblastoma (GBM) is the most aggressive malignant primary brain tumor characterized by a highly heterogeneous and immunosuppressive tumor microenvironment (TME). The symbiotic interactions between glioblastoma stem cells (GSCs) and tumor-associated macrophages (TAM) in the TME are critical for tumor progression. Here, we identified that IFI35, a transcriptional regulatory factor, plays both cell-intrinsic and cell-extrinsic roles in maintaining GSCs and the immunosuppressive TME. IFI35 induced non-canonical NF-kB signaling through proteasomal processing of p105 to the DNA-binding transcription factor p50, which heterodimerizes with RELB (RELB/p50), and activated cell chemotaxis in a cell-autonomous manner. Further, IFI35 induced recruitment and maintenance of M2-like TAMs in TME in a paracrine manner. Targeting IFI35 effectively suppressed in vivo tumor growth and prolonged survival of orthotopic xenograft-bearing mice. Collectively, these findings reveal the tumor-promoting functions of IFI35 and suggest that targeting IFI35 or its downstream effectors may provide effective approaches to improve GBM treatment.
© 2024. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

