Leucoma salicis nucleopolyhedrovirus (LesaNPV) genome sequence shed new light on the origin of the Alphabaculovirus orpseudotsugatae species
- PMID: 38594489
- PMCID: PMC11139710
- DOI: 10.1007/s11262-024-02062-x
Leucoma salicis nucleopolyhedrovirus (LesaNPV) genome sequence shed new light on the origin of the Alphabaculovirus orpseudotsugatae species
Abstract
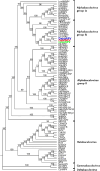
LesaNPV (Leucoma salicis nucleopolyhedrovirus) is an alphabaculovirus group Ib. Potentially, it can be an eco-friendly agent to control the white satin moth Leucoma salicis population. In this study, we have established the relationship between LesaNPV and other closely related alphabaculoviruses. Environmental samples of late instar of white satin moth collected in Poland infected with baculovirus have been homogenized, polyhedra were purified and subjected to scanning and transmission electron microscopy. Viral DNA was sequenced using the Illumina platform and the whole-genome sequence was established by de novo assembly of paired reads. Genome annotation and phylogenetic analyses were performed with the use of bioinformatics tools. The genome of LesaNPV is 132 549 bp long with 154 ORFs and 54.9% GC content. Whole-genome sequencing revealed deletion of dUTPase as well as ribonucleoside reductases small and large subunits region in LesaNPV genome compared to Dasychira pudibunda nucleopolyhedrovirus (DapuNPV) and Orgyia pseudotsugata multiple nucleopolyhedrovirus (OpMNPV) where this region is complete. Phylogenetic analysis of Baculoviridae family members showed that LesaNPV is less divergent from a common ancestor than closely related species DapuNPV and OpMNPV. This is interesting because their hosts do not occur in the same area. The baculoviruses described in this manuscript are probably isolates of one species and could be assigned to recently denominated species Alphabaculovirus orpseudotsugatae, historically originating from OpMNPV. This finding could have significant implications for the classification and understanding of the phylogeographical spread of baculoviruses.
Keywords: Leucoma salicis; Alphabaculovirus; Common ancestor; DapuNPV; LesaNPV; OpMNPV.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
The genome of Dasychira pudibunda nucleopolyhedrovirus (DapuNPV) reveals novel genetic connection between baculoviruses infecting moths of the Lymantriidae family.BMC Genomics. 2015 Oct 8;16:759. doi: 10.1186/s12864-015-1963-9. BMC Genomics. 2015. PMID: 26449402 Free PMC article.
-
Complete genome sequence analysis and genome organization of Dasychira pudibunda nucleopolyhedrovirus (DapuNPV-T1) from Turkey.Arch Microbiol. 2023 Dec 11;206(1):16. doi: 10.1007/s00203-023-03741-3. Arch Microbiol. 2023. PMID: 38079009
-
A Nymphalid-Infecting Group I Alphabaculovirus Isolated from the Major Passion Fruit Caterpillar Pest Dione juno juno (Lepidoptera: Nymphalidae).Viruses. 2019 Jul 3;11(7):602. doi: 10.3390/v11070602. Viruses. 2019. PMID: 31277203 Free PMC article.
-
European Leucoma salicis NPV is closely related to North American Orgyia pseudotsugata MNPV.J Invertebr Pathol. 2005 Feb;88(2):100-7. doi: 10.1016/j.jip.2004.12.002. J Invertebr Pathol. 2005. PMID: 15766926
-
Complete sequence, analysis and organization of the Orgyia leucostigma nucleopolyhedrovirus genome.Viruses. 2011 Nov;3(11):2301-27. doi: 10.3390/v3112301. Epub 2011 Nov 15. Viruses. 2011. PMID: 22163346 Free PMC article.
References
-
- Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Davison AJ, Dempsey DM, Dutilh BE, García ML, Harrach B, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Krupovic M, Kuhn JH, Lambert AJ, Łobocka M, Nibert ML, Oksanen HM, Orton RJ, Robertson DL, Rubino L, Sabanadzovic S, Simmonds P, Smith DB, Suzuki N, Van Dooerslaer K, Vandamme A-M, Varsani A, Zerbini FM. Changes to virus taxonomy and to the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2021) Arch Virol. 2021;166:2633–2648. doi: 10.1007/s00705-021-05156-1. - DOI - PubMed
-
- Javed MA, Biswas S, Willis LG, Harris S, Pritchard C, van Oers MM, Donly BC, Erlandson MA, Hegedus DD, Theilmann DA. Autographa californica multiple nucleopolyhedrovirus AC83 is a per os infectivity factor (PIF) protein required for occlusion-derived virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the PIF complex in ODV envelopes. J Virol. 2017;91:e02115–e2116. doi: 10.1128/JVI.02115-16. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

