CD32 captures committed haemogenic endothelial cells during human embryonic development
- PMID: 38594587
- PMCID: PMC11098737
- DOI: 10.1038/s41556-024-01403-0
CD32 captures committed haemogenic endothelial cells during human embryonic development
Abstract
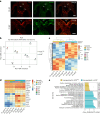
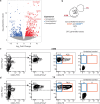
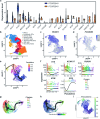
During embryonic development, blood cells emerge from specialized endothelial cells, named haemogenic endothelial cells (HECs). As HECs are rare and only transiently found in early developing embryos, it remains difficult to distinguish them from endothelial cells. Here we performed transcriptomic analysis of 28- to 32-day human embryos and observed that the expression of Fc receptor CD32 (FCGR2B) is highly enriched in the endothelial cell population that contains HECs. Functional analyses using human embryonic and human pluripotent stem cell-derived endothelial cells revealed that robust multilineage haematopoietic potential is harboured within CD32+ endothelial cells and showed that 90% of CD32+ endothelial cells are bona fide HECs. Remarkably, these analyses indicated that HECs progress through different states, culminating in FCGR2B expression, at which point cells are irreversibly committed to a haematopoietic fate. These findings provide a precise method for isolating HECs from human embryos and human pluripotent stem cell cultures, thus allowing the efficient generation of haematopoietic cells in vitro.
© 2024. The Author(s).
Conflict of interest statement
A.D. and R.S. have filed a patent application for the methodology described in this manuscript. C.M.S. is a scientific founder and Scientific Advisory Board member of Clade Therapeutics. The other authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
- ANR-14-CE11-0008/Agence Nationale de la Recherche (French National Research Agency)
- 101044032/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- R01HL151777/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- 26779-2021/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- R01 HL145290/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

