Cytosolic DNA sensor AIM2 promotes KRAS-driven lung cancer independent of inflammasomes
- PMID: 38594840
- PMCID: PMC11145135
- DOI: 10.1111/cas.16171
Cytosolic DNA sensor AIM2 promotes KRAS-driven lung cancer independent of inflammasomes
Abstract
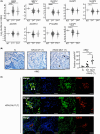
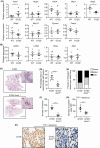
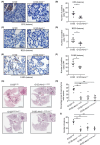
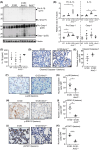
Constitutively active KRAS mutations are among the major drivers of lung cancer, yet the identity of molecular co-operators of oncogenic KRAS in the lung remains ill-defined. The innate immune cytosolic DNA sensor and pattern recognition receptor (PRR) Absent-in-melanoma 2 (AIM2) is best known for its assembly of multiprotein inflammasome complexes and promoting an inflammatory response. Here, we define a role for AIM2, independent of inflammasomes, in KRAS-addicted lung adenocarcinoma (LAC). In genetically defined and experimentally induced (nicotine-derived nitrosamine ketone; NNK) LAC mouse models harboring the KrasG12D driver mutation, AIM2 was highly upregulated compared with other cytosolic DNA sensors and inflammasome-associated PRRs. Genetic ablation of AIM2 in KrasG12D and NNK-induced LAC mouse models significantly reduced tumor growth, coincident with reduced cellular proliferation in the lung. Bone marrow chimeras suggest a requirement for AIM2 in KrasG12D-driven LAC in both hematopoietic (immune) and non-hematopoietic (epithelial) cellular compartments, which is supported by upregulated AIM2 expression in immune and epithelial cells of mutant KRAS lung tissues. Notably, protection against LAC in AIM2-deficient mice is associated with unaltered protein levels of mature Caspase-1 and IL-1β inflammasome effectors. Moreover, genetic ablation of the key inflammasome adapter, ASC, did not suppress KrasG12D-driven LAC. In support of these in vivo findings, AIM2, but not mature Caspase-1, was upregulated in human LAC patient tumor biopsies. Collectively, our findings reveal that endogenous AIM2 plays a tumor-promoting role, independent of inflammasomes, in mutant KRAS-addicted LAC, and suggest innate immune DNA sensing may provide an avenue to explore new therapeutic strategies in lung cancer.
Keywords: cell proliferation; inflammasome; innate immunity; lung cancer; pattern recognition receptors.
© 2024 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
The author declares no conflict of interest.
Figures


References
-
- Ahrendt SA, Decker PA, Alawi EA, et al. Cigarette smoking is strongly associated with mutation of the K‐ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92(6):1525‐1530. - PubMed
-
- Meng D, Yuan M, Li X, et al. Prognostic value of K‐RAS mutations in patients with non‐small cell lung cancer: a systematic review with meta‐analysis. Lung Cancer. 2013;81(1):1‐10. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

