Prostate cancer research: tools, cell types, and molecular targets
- PMID: 38595814
- PMCID: PMC11002103
- DOI: 10.3389/fonc.2024.1321694
Prostate cancer research: tools, cell types, and molecular targets
Abstract
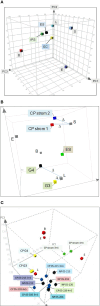
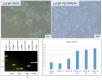
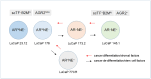
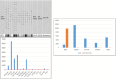
Multiple cancer cell types are found in prostate tumors. They are either luminal-like adenocarcinoma or less luminal-like and more stem-like non-adenocarcinoma and small cell carcinoma. These types are lineage related through differentiation. Loss of cancer differentiation from luminal-like to stem-like is mediated by the activation of stem cell transcription factors (scTF) such as LIN28A, NANOG, POU5F1 and SOX2. scTF expression leads to down-regulation of β2-microglobulin (B2M). Thus, cancer cells can change from the phenotype of differentiated to that of of dedifferentiated in the disease course. In development, epithelial cell differentiation is induced by stromal signaling and cell contact. One of the stromal factors specific to prostate encodes proenkephalin (PENK). PENK can down-regulate scTF and up-regulate B2M in stem-like small cell carcinoma LuCaP 145.1 cells indicative of exit from the stem state and differentiation. In fact, prostate cancer cells can be made to undergo dedifferentiation or reprogramming by scTF transfection and then to differentiate by PENK transfection. Therapies need to be designed for treating the different cancer cell types. Extracellular anterior gradient 2 (eAGR2) is an adenocarcinoma antigen associated with cancer differentiation that can be targeted by antibodies to lyse tumor cells with immune system components. eAGR2 is specific to cancer as normal cells express only the intracellular form (iAGR2). For AGR2-negative stem-like cancer cells, factors like PENK that can target scTF could be effective in differentiation therapy.
Keywords: AGR2 immunotherapy; cancer cell reprogramming; cancer differentiation; differentiation therapy; lineage relationship; stem cell transcription factors; stromal PENK.
Copyright © 2024 Liu.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
The opposing action of stromal cell proenkephalin and stem cell transcription factors in prostate cancer differentiation.BMC Cancer. 2021 Dec 15;21(1):1335. doi: 10.1186/s12885-021-09090-y. BMC Cancer. 2021. PMID: 34911496 Free PMC article.
-
Lineage relationship between prostate adenocarcinoma and small cell carcinoma.BMC Cancer. 2019 May 30;19(1):518. doi: 10.1186/s12885-019-5680-7. BMC Cancer. 2019. PMID: 31146720 Free PMC article.
-
Conversion of Prostate Adenocarcinoma to Small Cell Carcinoma-Like by Reprogramming.J Cell Physiol. 2016 Sep;231(9):2040-7. doi: 10.1002/jcp.25313. Epub 2016 Feb 4. J Cell Physiol. 2016. PMID: 26773436
-
Differentiation pathways and histogenetic aspects of normal and abnormal prostatic growth: a stem cell model.Prostate. 1996 Feb;28(2):98-106. doi: 10.1002/(SICI)1097-0045(199602)28:2<98::AID-PROS4>3.0.CO;2-J. Prostate. 1996. PMID: 8604398 Review.
-
Maturation arrest of stem cell differentiation is a common pathway for the cellular origin of teratocarcinomas and epithelial cancers.Lab Invest. 1994 Jan;70(1):6-22. Lab Invest. 1994. PMID: 8302019 Review.
Cited by
-
Interpretable machine learning driven biomarker identification and validation for prostate cancer.Transl Androl Urol. 2025 Jun 30;14(6):1528-1541. doi: 10.21037/tau-2025-242. Epub 2025 Jun 26. Transl Androl Urol. 2025. PMID: 40687654 Free PMC article.
-
Prostate cancer stem cells: an updated mini-review.J Cancer. 2024 Oct 28;15(20):6570-6576. doi: 10.7150/jca.100604. eCollection 2024. J Cancer. 2024. PMID: 39668824 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

