Cat-E: A comprehensive web tool for exploring cancer targeting strategies
- PMID: 38596315
- PMCID: PMC11001601
- DOI: 10.1016/j.csbj.2024.03.024
Cat-E: A comprehensive web tool for exploring cancer targeting strategies
Abstract
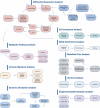
Identifying potential cancer-associated genes and drug targets from omics data is challenging due to its diverse sources and analyses, requiring advanced skills and large amounts of time. To facilitate such analysis, we developed Cat-E (Cancer Target Explorer), a novel R/Shiny web tool designed for comprehensive analysis with evaluation according to cancer-related omics data. Cat-E is accessible at https://cat-e.bioinfo-wuerz.eu/. Cat-E compiles information on oncolytic viruses, cell lines, gene markers, and clinical studies by integrating molecular datasets from key databases such as OvirusTB, TCGA, DrugBANK, and PubChem. Users can use all datasets and upload their data to perform multiple analyses, such as differential gene expression analysis, metabolic pathway exploration, metabolic flux analysis, GO and KEGG enrichment analysis, survival analysis, immune signature analysis, single nucleotide variation analysis, dynamic analysis of gene expression changes and gene regulatory network changes, and protein structure prediction. Cancer target evaluation by Cat-E is demonstrated here on lung adenocarcinoma (LUAD) datasets. By offering a user-friendly interface and detailed user manual, Cat-E eliminates the need for advanced computational expertise, making it accessible to experimental biologists, undergraduate and graduate students, and oncology clinicians. It serves as a valuable tool for investigating genetic variations across diverse cancer types, facilitating the identification of novel diagnostic markers and potential therapeutic targets.
Keywords: Cancer; Cancer pathways; Immune modulation; Network analysis; Oncolytic virus; Protein interaction.
© 2024 The Authors.
Figures






Similar articles
-
IPAD: the Integrated Pathway Analysis Database for Systematic Enrichment Analysis.BMC Bioinformatics. 2012;13 Suppl 15(Suppl 15):S7. doi: 10.1186/1471-2105-13-S15-S7. Epub 2012 Sep 11. BMC Bioinformatics. 2012. PMID: 23046449 Free PMC article.
-
GENAVi: a shiny web application for gene expression normalization, analysis and visualization.BMC Genomics. 2019 Oct 16;20(1):745. doi: 10.1186/s12864-019-6073-7. BMC Genomics. 2019. PMID: 31619158 Free PMC article.
-
Clinical Significance and Immunometabolism Landscapes of a Novel Recurrence-Associated Lipid Metabolism Signature In Early-Stage Lung Adenocarcinoma: A Comprehensive Analysis.Front Immunol. 2022 Feb 10;13:783495. doi: 10.3389/fimmu.2022.783495. eCollection 2022. Front Immunol. 2022. PMID: 35222371 Free PMC article.
-
Leveraging diverse cell-death patterns to predict the clinical outcome of immune checkpoint therapy in lung adenocarcinoma: Based on muti-omics analysis and vitro assay.Oncol Res. 2023 Dec 28;32(2):393-407. doi: 10.32604/or.2023.031134. eCollection 2023. Oncol Res. 2023. PMID: 38186574 Free PMC article.
-
The Cancer Omics Atlas: an integrative resource for cancer omics annotations.BMC Med Genomics. 2018 Aug 8;11(1):63. doi: 10.1186/s12920-018-0381-7. BMC Med Genomics. 2018. PMID: 30089500 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
