Differences in F pocket impact on HLA I genetic associations with autoimmune diabetes
- PMID: 38596688
- PMCID: PMC11003304
- DOI: 10.3389/fimmu.2024.1342335
Differences in F pocket impact on HLA I genetic associations with autoimmune diabetes
Abstract
Introduction: Human leukocyte antigen (HLA) I molecules present antigenic peptides to activate CD8+ T cells. Type 1 Diabetes (T1D) is an auto-immune disease caused by aberrant activation of the CD8+ T cells that destroy insulin-producing pancreatic β cells. Some HLA I alleles were shown to increase the risk of T1D (T1D-predisposing alleles), while some reduce this risk (T1D-protective alleles).
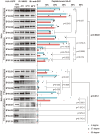
Methods: Here, we compared the T1D-predisposing and T1D-protective allotypes concerning peptide binding, maturation, localization and surface expression and correlated it with their sequences and energetic profiles using experimental and computational methods.
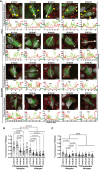
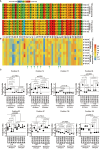
Results: T1D-predisposing allotypes had more peptide-bound forms and higher plasma membrane levels than T1D-protective allotypes. This was related to the fact that position 116 within the F pocket was more conserved and made more optimal contacts with the neighboring residues in T1D-predisposing allotypes than in protective allotypes.
Conclusion: Our work uncovers that specific polymorphisms in HLA I molecules potentially influence their susceptibility to T1D.
Keywords: HLA class I; antigen presentation; autoimmune diabetes; inflammation; peptide binding; polymorphism.
Copyright © 2024 Ren, Amarajeewa, Jayasinghe and Garstka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Low HLA binding of diabetes-associated CD8+ T-cell epitopes is increased by post translational modifications.BMC Immunol. 2018 Mar 21;19(1):12. doi: 10.1186/s12865-018-0250-3. BMC Immunol. 2018. PMID: 29562882 Free PMC article.
-
Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.J Biol Chem. 2024 Sep;300(9):107702. doi: 10.1016/j.jbc.2024.107702. Epub 2024 Aug 22. J Biol Chem. 2024. PMID: 39173948 Free PMC article.
-
Polymorphism in F pocket affects peptide selection and stability of type 1 diabetes-associated HLA-B39 allotypes.Eur J Immunol. 2024 Jun;54(6):e2350683. doi: 10.1002/eji.202350683. Epub 2024 Mar 29. Eur J Immunol. 2024. PMID: 38549458
-
Prediction of the risk of type 1 diabetes from polymorphisms in candidate genes.Diabetes Res Clin Pract. 2004 Dec;66 Suppl 1:S19-25. doi: 10.1016/j.diabres.2003.10.026. Diabetes Res Clin Pract. 2004. PMID: 15563974 Review.
-
Bridging Mice to Men: Using HLA Transgenic Mice to Enhance the Future Prediction and Prevention of Autoimmune Type 1 Diabetes in Humans.Methods Mol Biol. 2016;1438:137-51. doi: 10.1007/978-1-4939-3661-8_9. Methods Mol Biol. 2016. PMID: 27150089 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

