RNA and neuronal function: the importance of post-transcriptional regulation
- PMID: 38596700
- PMCID: PMC10913846
- DOI: 10.1093/oons/kvac011
RNA and neuronal function: the importance of post-transcriptional regulation
Abstract
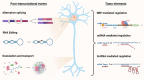
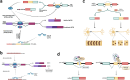
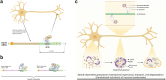
The brain represents an organ with a particularly high diversity of genes that undergo post-transcriptional gene regulation through multiple mechanisms that affect RNA metabolism and, consequently, brain function. This vast regulatory process in the brain allows for a tight spatiotemporal control over protein expression, a necessary factor due to the unique morphologies of neurons. The numerous mechanisms of post-transcriptional regulation or translational control of gene expression in the brain include alternative splicing, RNA editing, mRNA stability and transport. A large number of trans-elements such as RNA-binding proteins and micro RNAs bind to specific cis-elements on transcripts to dictate the fate of mRNAs including its stability, localization, activation and degradation. Several trans-elements are exemplary regulators of translation, employing multiple cofactors and regulatory machinery so as to influence mRNA fate. Networks of regulatory trans-elements exert control over key neuronal processes such as neurogenesis, synaptic transmission and plasticity. Perturbations in these networks may directly or indirectly cause neuropsychiatric and neurodegenerative disorders. We will be reviewing multiple mechanisms of gene regulation by trans-elements occurring specifically in neurons.
Keywords: RNA editing and neuronal granules; alternative splicing.
© The Author(s) 2022. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


References
-
- Aliaga EE, Mendoza I, Tapia-Arancibia L. Distinct subcellular localization of BDNF transcripts in cultured hypothalamic neurons and modification by neuronal activation. J Neural Transm (Vienna). 2009;116:23–32 - PubMed
-
- Andreadis A, Gallego ME, Nadal-Ginard B. Generation of protein isoform diversity by alternative splicing: mechanistic and biological implications. Annu Rev Cell Biol. 1987;3:207–42 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
