HELLS regulates transcription in T-cell lymphomas by reducing unscheduled R-loops and by facilitating RNAPII progression
- PMID: 38597676
- PMCID: PMC11194065
- DOI: 10.1093/nar/gkae239
HELLS regulates transcription in T-cell lymphomas by reducing unscheduled R-loops and by facilitating RNAPII progression
Abstract
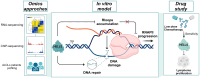
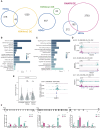
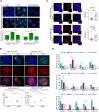
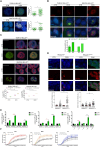
Chromatin modifiers are emerging as major determinants of many types of cancers, including Anaplastic Large Cell Lymphomas (ALCL), a family of highly heterogeneous T-cell lymphomas for which therapeutic options are still limited. HELLS is a multifunctional chromatin remodeling protein that affects genomic instability by participating in the DNA damage response. Although the transcriptional function of HELLS has been suggested, no clues on how HELLS controls transcription are currently available. In this study, by integrating different multi-omics and functional approaches, we characterized the transcriptional landscape of HELLS in ALCL. We explored the clinical impact of its transcriptional program in a large cohort of 44 patients with ALCL. We demonstrated that HELLS, loaded at the level of intronic regions of target promoters, facilitates RNA Polymerase II (RNAPII) progression along the gene bodies by reducing the persistence of co-transcriptional R-loops and promoting DNA damage resolution. Importantly, selective knockdown of HELLS sensitizes ALCL cells to different chemotherapeutic agents, showing a synergistic effect. Collectively, our work unveils the role of HELLS in acting as a gatekeeper of ALCL genome stability providing a rationale for drug design.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Fiore D., Cappelli L.V., Broccoli A., Zinzani P.L., Chan W.C., Inghirami G. Peripheral T cell lymphomas: from the bench to the clinic. Nat. Rev. Cancer. 2020; 20:323–342. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

