NetMe 2.0: a web-based platform for extracting and modeling knowledge from biomedical literature as a labeled graph
- PMID: 38597890
- PMCID: PMC11074003
- DOI: 10.1093/bioinformatics/btae194
NetMe 2.0: a web-based platform for extracting and modeling knowledge from biomedical literature as a labeled graph
Abstract
Motivation: The rapid increase of bio-medical literature makes it harder and harder for scientists to keep pace with the discoveries on which they build their studies. Therefore, computational tools have become more widespread, among which network analysis plays a crucial role in several life-science contexts. Nevertheless, building correct and complete networks about some user-defined biomedical topics on top of the available literature is still challenging.
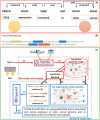
Results: We introduce NetMe 2.0, a web-based platform that automatically extracts relevant biomedical entities and their relations from a set of input texts-i.e. in the form of full-text or abstract of PubMed Central's papers, free texts, or PDFs uploaded by users-and models them as a BioMedical Knowledge Graph (BKG). NetMe 2.0 also implements an innovative Retrieval Augmented Generation module (Graph-RAG) that works on top of the relationships modeled by the BKG and allows the distilling of well-formed sentences that explain their content. The experimental results show that NetMe 2.0 can infer comprehensive and reliable biological networks with significant Precision-Recall metrics when compared to state-of-the-art approaches.
Availability and implementation: https://netme.click/.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
Similar articles
-
NETME: on-the-fly knowledge network construction from biomedical literature.Appl Netw Sci. 2022;7(1):1. doi: 10.1007/s41109-021-00435-x. Epub 2022 Jan 6. Appl Netw Sci. 2022. PMID: 35013714 Free PMC article.
-
ANDDigest: a new web-based module of ANDSystem for the search of knowledge in the scientific literature.BMC Bioinformatics. 2020 Sep 14;21(Suppl 11):228. doi: 10.1186/s12859-020-03557-8. BMC Bioinformatics. 2020. PMID: 32921303 Free PMC article.
-
Text mining-based word representations for biomedical data analysis and protein-protein interaction networks in machine learning tasks.PLoS One. 2021 Oct 15;16(10):e0258623. doi: 10.1371/journal.pone.0258623. eCollection 2021. PLoS One. 2021. PMID: 34653224 Free PMC article.
-
Text Mining for Building Biomedical Networks Using Cancer as a Case Study.Biomolecules. 2021 Sep 29;11(10):1430. doi: 10.3390/biom11101430. Biomolecules. 2021. PMID: 34680062 Free PMC article. Review.
-
PubMed and beyond: a survey of web tools for searching biomedical literature.Database (Oxford). 2011 Jan 18;2011:baq036. doi: 10.1093/database/baq036. Print 2011. Database (Oxford). 2011. PMID: 21245076 Free PMC article. Review.
Cited by
-
BioGSF: a graph-driven semantic feature integration framework for biomedical relation extraction.Brief Bioinform. 2024 Nov 22;26(1):bbaf025. doi: 10.1093/bib/bbaf025. Brief Bioinform. 2024. PMID: 39853110 Free PMC article.
-
CovidTGI: A tool to investigate the temporal genetic instability of SARS-CoV-2 variants.iScience. 2025 Mar 28;28(4):112315. doi: 10.1016/j.isci.2025.112315. eCollection 2025 Apr 18. iScience. 2025. PMID: 40264800 Free PMC article.
-
Darling (v2.0): Mining disease-related databases for the detection of biomedical entity associations.Comput Struct Biotechnol J. 2025 Jun 14;27:2626-2637. doi: 10.1016/j.csbj.2025.06.025. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40599243 Free PMC article.
References
-
- Beck J. Report from the field: Pubmed central, an xml-based archive of life sciences journal articles. In: Proceedings of the International Symposium on XML for the Long Haul: Issues in the Long-term Preservation of XML. Balisage Series on Markup Technologies, Montréal, Canada. Mulberry Technologies, Inc. Vol. 6 2010.
-
- Cai D, Wang Y, Liu L. et al. Recent advances in retrieval-augmented text generation. In: Proceedings of the 45th ACM SIGIR Conference, SIGIR ’22, New York NY United States, 2022, 3417–9.

