Bacterial growth dynamics in a rhythmic symbiosis
- PMID: 38598294
- PMCID: PMC11238090
- DOI: 10.1091/mbc.E24-01-0044
Bacterial growth dynamics in a rhythmic symbiosis
Abstract
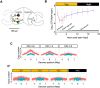
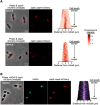
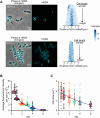
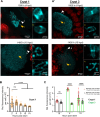
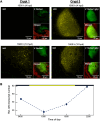
The symbiotic relationship between the bioluminescent bacterium Vibrio fischeri and the bobtail squid Euprymna scolopes serves as a valuable system to investigate bacterial growth and peptidoglycan (PG) synthesis within animal tissues. To better understand the growth dynamics of V. fischeri in the crypts of the light-emitting organ of its juvenile host, we showed that, after the daily dawn-triggered expulsion of most of the population, the remaining symbionts rapidly proliferate for ∼6 h. At that point the population enters a period of extremely slow growth that continues throughout the night until the next dawn. Further, we found that PG synthesis by the symbionts decreases as they enter the slow-growing stage. Surprisingly, in contrast to the most mature crypts (i.e., Crypt 1) of juvenile animals, most of the symbiont cells in the least mature crypts (i.e., Crypt 3) were not expelled and, instead, remained in the slow-growing state throughout the day, with almost no cell division. Consistent with this observation, the expression of the gene encoding the PG-remodeling enzyme, L,D-transpeptidase (LdtA), was greatest during the slowly growing stage of Crypt 1 but, in contrast, remained continuously high in Crypt 3. Finally, deletion of the ldtA gene resulted in a symbiont that grew and survived normally in culture, but was increasingly defective in competing against its parent strain in the crypts. This result suggests that remodeling of the PG to generate additional 3-3 linkages contributes to the bacterium's fitness in the symbiosis, possibly in response to stresses encountered during the very slow-growing stage.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

