The bistable mitotic switch in fission yeast
- PMID: 38598296
- PMCID: PMC11238088
- DOI: 10.1091/mbc.E24-03-0142
The bistable mitotic switch in fission yeast
Abstract
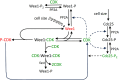
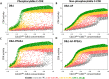
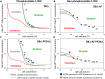
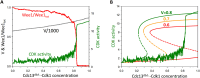
In favorable conditions, eukaryotic cells proceed irreversibly through the cell division cycle (G1-S-G2-M) in order to produce two daughter cells with the same number and identity of chromosomes of their progenitor. The integrity of this process is maintained by "checkpoints" that hold a cell at particular transition points of the cycle until all requisite events are completed. The crucial functions of these checkpoints seem to depend on irreversible bistability of the underlying checkpoint control systems. Bistability of cell cycle transitions has been confirmed experimentally in frog egg extracts, budding yeast cells and mammalian cells. For fission yeast cells, a recent paper by Patterson et al. (2021) provides experimental evidence for an abrupt transition from G2 phase into mitosis, and we show that these data are consistent with a stochastic model of a bistable switch governing the G2/M checkpoint. Interestingly, our model suggests that their experimental data could also be explained by a reversible/sigmoidal switch, and stochastic simulations confirm this supposition. We propose a simple modification of their experimental protocol that could provide convincing evidence for (or against) bistability of the G2/M transition in fission yeast.
Figures




Similar articles
-
Mathematical model of the fission yeast cell cycle with checkpoint controls at the G1/S, G2/M and metaphase/anaphase transitions.Biophys Chem. 1998 May 5;72(1-2):185-200. doi: 10.1016/s0301-4622(98)00133-1. Biophys Chem. 1998. PMID: 9652094
-
How do fission yeast cells grow and connect growth to the mitotic cycle?Curr Genet. 2017 May;63(2):165-173. doi: 10.1007/s00294-016-0632-0. Epub 2016 Jul 27. Curr Genet. 2017. PMID: 27465359 Review.
-
The chk1 pathway is required to prevent mitosis following cell-cycle arrest at 'start'.Curr Biol. 1995 Oct 1;5(10):1179-90. doi: 10.1016/s0960-9822(95)00234-x. Curr Biol. 1995. PMID: 8548290
-
The kinetics of the B cyclin p56cdc13 and the phosphatase p80cdc25 during the cell cycle of the fission yeast Schizosaccharomyces pombe.J Cell Sci. 1996 Jun;109 ( Pt 6):1647-53. doi: 10.1242/jcs.109.6.1647. J Cell Sci. 1996. PMID: 8799851
-
HIV-1 VPR modulates cell cycle G2/M transition through an alternative cellular mechanism other than the classic mitotic checkpoints.Front Biosci. 2002 Feb 1;7:d349-57. doi: 10.2741/elder. Front Biosci. 2002. PMID: 11815283 Review.
References
-
- Acar M, Becskei A, van Oudenaarden A (2005). Enhancement of cellular memory by reducing stochastic transitions. Nature 435, 228–232. - PubMed
-
- Chica N, Rozalen AE, Perez-Hidalgo L, Rubio A, Novak B, Moreno S (2016). Nutritional Control of Cell Size by the Greatwall-Endosulfine-PP2A.B55 Pathway. Curr Biol 26, 319–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

