The proliferation of antibiotic resistance genes (ARGs) and microbial communities in industrial wastewater treatment plant treating N,N-dimethylformamide (DMF) by AAO process
- PMID: 38598457
- PMCID: PMC11006197
- DOI: 10.1371/journal.pone.0299740
The proliferation of antibiotic resistance genes (ARGs) and microbial communities in industrial wastewater treatment plant treating N,N-dimethylformamide (DMF) by AAO process
Abstract
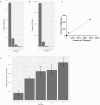
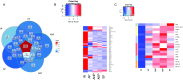
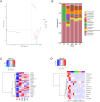
The excessive use of antibiotics has resulted in the contamination of the environment with antibiotic resistance genes (ARGs), posing a significant threat to public health. Wastewater treatment plants (WWTPs) are known to be reservoirs of ARGs and considered to be hotspots for horizontal gene transfer (HGT) between bacterial communities. However, most studies focused on the distribution and dissemination of ARGs in hospital and urban WWTPs, and little is known about their fate in industrial WWTPs. In this study, collected the 15 wastewater samples containing N,N-dimethylformamide (DMF) from five stages of the anaerobic anoxic aerobic (AAO) process in an industrial WWTPs. The findings revealed a stepwise decrease in DMF and chemical oxygen demand (COD) content with the progression of treatment. However, the number and abundances of ARGs increase in the effluents of biological treatments. Furthermore, the residues of DMF and the treatment process altered the structure of the bacterial community. The correlation analysis indicated that the shift in bacterial community structures might be the main driver for the dynamics change of ARGs. Interestingly, observed that the AAO process may acted as a microbial source and increased the total abundance of ARGs instead of attenuating it. Additionally, found that non-pathogenic bacteria had higher ARGs abundance than pathogenic bacteria in effluents. The study provides insights into the microbial community structure and the mechanisms that drive the variation in ARGs abundance in industrial WWTPs.
Copyright: © 2024 Gao et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Variations in antibiotic resistance genes and microbial community in sludges passing through biological nutrient removal and anaerobic digestion processes in municipal wastewater treatment plants.Chemosphere. 2023 Feb;313:137362. doi: 10.1016/j.chemosphere.2022.137362. Epub 2022 Nov 22. Chemosphere. 2023. PMID: 36427585
-
Characterisation of microbial communities and quantification of antibiotic resistance genes in Italian wastewater treatment plants using 16S rRNA sequencing and digital PCR.Sci Total Environ. 2024 Jul 10;933:173217. doi: 10.1016/j.scitotenv.2024.173217. Epub 2024 May 13. Sci Total Environ. 2024. PMID: 38750766
-
Unveiling dynamics of size-dependent antibiotic resistome associated with microbial communities in full-scale wastewater treatment plants.Water Res. 2020 Dec 15;187:116450. doi: 10.1016/j.watres.2020.116450. Epub 2020 Sep 23. Water Res. 2020. PMID: 32998097
-
Promising bioprocesses for the efficient removal of antibiotics and antibiotic-resistance genes from urban and hospital wastewaters: Potentialities of aerobic granular systems.Environ Pollut. 2024 Feb 1;342:123115. doi: 10.1016/j.envpol.2023.123115. Epub 2023 Dec 10. Environ Pollut. 2024. PMID: 38086508 Review.
-
Antibiotic resistant bacteria and genes in wastewater treatment plants: From occurrence to treatment strategies.Sci Total Environ. 2022 Sep 10;838(Pt 4):156544. doi: 10.1016/j.scitotenv.2022.156544. Epub 2022 Jun 6. Sci Total Environ. 2022. PMID: 35679932 Review.
Cited by
-
The surveillance of antimicrobial resistance in wastewater from a one health perspective: A global scoping and temporal review (2014-2024).One Health. 2025 Jul 14;21:101139. doi: 10.1016/j.onehlt.2025.101139. eCollection 2025 Dec. One Health. 2025. PMID: 40727444 Free PMC article.
References
-
- Riaz L, Wang Q, Yang Q, Li X, Yuan W. Potential of industrial composting and anaerobic digestion for the removal of antibiotics, antibiotic resistance genes and heavy metals from chicken manure. The Science of the total environment. 2020;718:137414. Epub 2020/02/28. doi: 10.1016/j.scitotenv.2020.137414 . - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

