Synovial fibroblast gene expression is associated with sensory nerve growth and pain in rheumatoid arthritis
- PMID: 38598614
- PMCID: PMC11931728
- DOI: 10.1126/scitranslmed.adk3506
Synovial fibroblast gene expression is associated with sensory nerve growth and pain in rheumatoid arthritis
Abstract
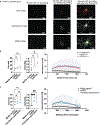
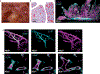
It has been presumed that rheumatoid arthritis (RA) joint pain is related to inflammation in the synovium; however, recent studies reveal that pain scores in patients do not correlate with synovial inflammation. We developed a machine-learning approach (graph-based gene expression module identification or GbGMI) to identify an 815-gene expression module associated with pain in synovial biopsy samples from patients with established RA who had limited synovial inflammation at arthroplasty. We then validated this finding in an independent cohort of synovial biopsy samples from patients who had early untreated RA with little inflammation. Single-cell RNA sequencing analyses indicated that most of these 815 genes were most robustly expressed by lining layer synovial fibroblasts. Receptor-ligand interaction analysis predicted cross-talk between human lining layer fibroblasts and human dorsal root ganglion neurons expressing calcitonin gene-related peptide (CGRP+). Both RA synovial fibroblast culture supernatant and netrin-4, which is abundantly expressed by lining fibroblasts and was within the GbGMI-identified pain-associated gene module, increased the branching of pain-sensitive murine CGRP+ dorsal root ganglion neurons in vitro. Imaging of solvent-cleared synovial tissue with little inflammation from humans with RA revealed CGRP+ pain-sensing neurons encasing blood vessels growing into synovial hypertrophic papilla. Together, these findings support a model whereby synovial lining fibroblasts express genes associated with pain that enhance the growth of pain-sensing neurons into regions of synovial hypertrophy in RA.
Conflict of interest statement
Figures
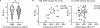




References
-
- Marchand F, Perretti M, McMahon SB, Role of the immune system in chronic pain. Nat. Rev. Neurosci. 6, 521–532 (2005). - PubMed
-
- Alivernini S, Firestein GS, McInnes IB, The pathogenesis of rheumatoid arthritis. Immunity 55, 2255–2270 (2022). - PubMed
-
- Buch MH, Eyre S, McGonagle D, Persistent inflammatory and non-inflammatory mechanisms in refractory rheumatoid arthritis. Nat. Rev. Rheumatol. 17, 17–33 (2021). - PubMed
-
- Nagy G, Roodenrijs NM, Welsing PM, Kedves M, Hamar A, van der Goes MC, Kent A, Bakkers M, Blaas E, Senolt L, Szekanecz Z, Choy E, Dougados M, Jacobs JW, Geenen R, Bijlsma HW, Zink A, Aletaha D, Schoneveld L, van Riel P, Gutermann L, Prior Y, Nikiphorou E, Ferraccioli G, Schett G, Hyrich KL, Mueller-Ladner U, Buch MH, McInnes IB, van der Heijde D, van Laar JM, EULAR definition of difficult-to-treat rheumatoid arthritis. Ann. Rheum. Dis. 80, 31–35 (2021). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UH2 AR067685/AR/NIAMS NIH HHS/United States
- UM2 AR067678/AR/NIAMS NIH HHS/United States
- R01 AR077019/AR/NIAMS NIH HHS/United States
- P30 AR079206/AR/NIAMS NIH HHS/United States
- R01 AR060364/AR/NIAMS NIH HHS/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- UH2 AR067688/AR/NIAMS NIH HHS/United States
- UH2 AR067689/AR/NIAMS NIH HHS/United States
- UH2 AR067690/AR/NIAMS NIH HHS/United States
- UH2 AR067677/AR/NIAMS NIH HHS/United States
- R01 AR078268/AR/NIAMS NIH HHS/United States
- UH2 AR067679/AR/NIAMS NIH HHS/United States
- UH2 AR067681/AR/NIAMS NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U19 NS130608/NS/NINDS NIH HHS/United States
- UC2 AR081025/AR/NIAMS NIH HHS/United States
- R01 AR064251/AR/NIAMS NIH HHS/United States
- UH2 AR067694/AR/NIAMS NIH HHS/United States
- UH2 AR067676/AR/NIAMS NIH HHS/United States
- UH2 AR067691/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

