A digital twin of the infant microbiome to predict neurodevelopmental deficits
- PMID: 38598636
- PMCID: PMC11006218
- DOI: 10.1126/sciadv.adj0400
A digital twin of the infant microbiome to predict neurodevelopmental deficits
Abstract
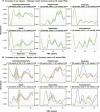
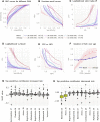
Despite the recognized gut-brain axis link, natural variations in microbial profiles between patients hinder definition of normal abundance ranges, confounding the impact of dysbiosis on infant neurodevelopment. We infer a digital twin of the infant microbiome, forecasting ecosystem trajectories from a few initial observations. Using 16S ribosomal RNA profiles from 88 preterm infants (398 fecal samples and 32,942 abundance estimates for 91 microbial classes), the model (Q-net) predicts abundance dynamics with R2 = 0.69. Contrasting the fit to Q-nets of typical versus suboptimal development, we can reliably estimate individual deficit risk (Mδ) and identify infants achieving poor future head circumference growth with ≈76% area under the receiver operator characteristic curve, 95% ± 1.8% positive predictive value at 98% specificity at 30 weeks postmenstrual age. We find that early transplantation might mitigate risk for ≈45.2% of the cohort, with potentially negative effects from incorrect supplementation. Q-nets are generative artificial intelligence models for ecosystem dynamics, with broad potential applications.
Figures




References
-
- Cryan J. F., Dinan T. G., Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712 (2012). - PubMed
-
- Baranowski J. R., Claud E. C., Necrotizing enterocolitis and the preterm infant microbiome. Adv. Exp. Med. Biol. 1125, 25–36 (2019). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

