Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel
- PMID: 38598639
- PMCID: PMC11006217
- DOI: 10.1126/sciadv.adl5952
Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel
Abstract
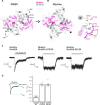
N-methyl-d-aspartate receptors (NMDARs) and other ionotropic glutamate receptors (iGluRs) mediate most of the excitatory signaling in the mammalian brains in response to the neurotransmitter glutamate. Uniquely, NMDARs composed of GluN1 and GluN3 are activated exclusively by glycine, the neurotransmitter conventionally mediating inhibitory signaling when it binds to pentameric glycine receptors. The GluN1-3 NMDARs are vital for regulating neuronal excitability, circuit function, and specific behaviors, yet our understanding of their functional mechanism at the molecular level has remained limited. Here, we present cryo-electron microscopy structures of GluN1-3A NMDARs bound to an antagonist, CNQX, and an agonist, glycine. The structures show a 1-3-1-3 subunit heterotetrameric arrangement and an unprecedented pattern of GluN3A subunit orientation shift between the glycine-bound and CNQX-bound structures. Site-directed disruption of the unique subunit interface in the glycine-bound structure mitigated desensitization. Our study provides a foundation for understanding the distinct structural dynamics of GluN3 that are linked to the unique function of GluN1-3 NMDARs.
Figures

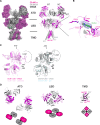



References
-
- Hansen K. B., Wollmuth L. P., Bowie D., Furukawa H., Menniti F. S., Sobolevsky A., Swanson G. T., Swanger S. A., Greger I. H., Nakagawa T., McBain C. J., Jayaraman V., Dell’Acqua M. L., Diamond J. S., Camp C. R., Perszyk R. E., Yuan H., Traynelis S., Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol. Rev. 73, 298–487 (2021). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

