Identifying the predictive role and the related drugs of oxidative stress genes in the hepatocellular carcinoma
- PMID: 38599581
- PMCID: PMC11006533
- DOI: 10.1002/cnr2.1978
Identifying the predictive role and the related drugs of oxidative stress genes in the hepatocellular carcinoma
Abstract
Background and aims: Oncogenesis and tumor development have been related to oxidative stress (OS). The potential diagnostic utility of OS genes in hepatocellular carcinoma (HCC), however, remains uncertain. As a result, this work aimed to create a novel OS related-genes signature that could be used to predict the survival of HCC patients and to screen OS related-genes drugs that might be used for HCC treatment.
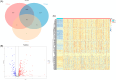
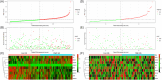
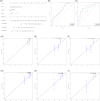
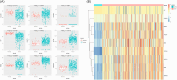
Methods: We used The Cancer Genome Atlas (TCGA) database and the Gene Expression Omnibus (GEO) database to acquire mRNA expression profiles and clinical data for this research and the GeneCards database to obtain OS related-genes. Following that, biological functions from Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed on differentially expressed OS-related genes (DEOSGs). Subsequently, the prognostic risk signature was constructed based on DEOSGs from the TCGA data that were screened by using univariate cox analysis, and the Least Absolute Shrinkage and Selection Operator (LASSO) regression, and multivariate cox analysis. At the same time, we developed a prognostic nomogram of HCC patients based on risk signature and clinical-pathological characteristics. The GEO data was used for validation. We used the receiver operating characteristic (ROC) curve, calibration curves, and Kaplan-Meier (KM) survival curves to examine the prediction value of the risk signature and nomogram. Finally, we screened the differentially expressed OS genes related drugs.
Results: We were able to recognize 9 OS genes linked to HCC prognosis. In addition, the KM curve revealed a statistically significant difference in overall survival (OS) between the high-risk and low-risk groups. The area under the curve (AUC) shows the independent prognostic value of the risk signature model. Meanwhile, the ROC curves and calibration curves show the strong prognostic power of the nomogram. The top three drugs with negative ratings were ZM-336372, lestaurtinib, and flunisolide, all of which inversely regulate different OS gene expressions.
Conclusion: Our findings indicate that OS related-genes have a favorable prognostic value for HCC, which sheds new light on the relationship between oxidative stress and HCC, and suggests potential therapeutic strategies for HCC patients.
Keywords: GEO; TCGA; hepatocellular carcinoma; overall survival; oxidative stress‐related genes; prognostic.
© 2024 The Authors. Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors state that no commercial or financial connections that might be considered as a possible conflict of interest existed during the research.
Figures





References
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

