Allopolyploid origin and diversification of the Hawaiian endemic mints
- PMID: 38600100
- PMCID: PMC11006916
- DOI: 10.1038/s41467-024-47247-y
Allopolyploid origin and diversification of the Hawaiian endemic mints
Abstract
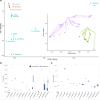
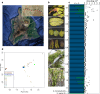
Island systems provide important contexts for studying processes underlying lineage migration, species diversification, and organismal extinction. The Hawaiian endemic mints (Lamiaceae family) are the second largest plant radiation on the isolated Hawaiian Islands. We generated a chromosome-scale reference genome for one Hawaiian species, Stenogyne calaminthoides, and resequenced 45 relatives, representing 34 species, to uncover the continental origins of this group and their subsequent diversification. We further resequenced 109 individuals of two Stenogyne species, and their purported hybrids, found high on the Mauna Kea volcano on the island of Hawai'i. The three distinct Hawaiian genera, Haplostachys, Phyllostegia, and Stenogyne, are nested inside a fourth genus, Stachys. We uncovered four independent polyploidy events within Stachys, including one allopolyploidy event underlying the Hawaiian mints and their direct western North American ancestors. While the Hawaiian taxa may have principally diversified by parapatry and drift in small and fragmented populations, localized admixture may have played an important role early in lineage diversification. Our genomic analyses provide a view into how organisms may have radiated on isolated island chains, settings that provided one of the principal natural laboratories for Darwin's thinking about the evolutionary process.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Stearns, H. T. Geology of the Hawaiian islands. (Honolulu Advertiser, 1946).
-
- Stone, C. P. & Stone, D. B. Conservation biology in Hawaiʻi, (University of Hawaii Cooperative National Park Resources Studies Unit, Honolulu, Hawaiʻi, 1989).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

