Endogenous aldehyde-induced DNA-protein crosslinks are resolved by transcription-coupled repair
- PMID: 38600234
- PMCID: PMC11098742
- DOI: 10.1038/s41556-024-01401-2
Endogenous aldehyde-induced DNA-protein crosslinks are resolved by transcription-coupled repair
Abstract
DNA-protein crosslinks (DPCs) induced by aldehydes interfere with replication and transcription. Hereditary deficiencies in DPC repair and aldehyde clearance processes cause progeria, including Ruijs-Aalfs syndrome (RJALS) and AMeD syndrome (AMeDS) in humans. Although the elimination of DPC during replication has been well established, how cells overcome DPC lesions in transcription remains elusive. Here we show that endogenous aldehyde-induced DPC roadblocks are efficiently resolved by transcription-coupled repair (TCR). We develop a high-throughput sequencing technique to measure the genome-wide distribution of DPCs (DPC-seq). Using proteomics and DPC-seq, we demonstrate that the conventional TCR complex as well as VCP/p97 and the proteasome are required for the removal of formaldehyde-induced DPCs. TFIIS-dependent cleavage of RNAPII transcripts protects against transcription obstacles. Finally, a mouse model lacking both aldehyde clearance and TCR confirms endogenous DPC accumulation in actively transcribed regions. Collectively, our data provide evidence that transcription-coupled DPC repair (TC-DPCR) as well as aldehyde clearance are crucial for protecting against metabolic genotoxin, thus explaining the molecular pathogenesis of AMeDS and other disorders associated with defects in TCR, such as Cockayne syndrome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
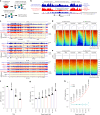


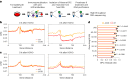
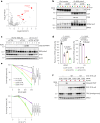




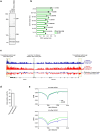
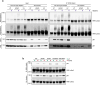
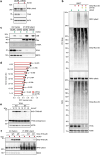

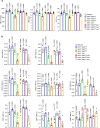

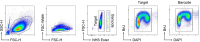
References
Publication types
MeSH terms
Grants and funding
- JP21ek0109549/Japan Agency for Medical Research and Development (AMED)
- JP22ek0109559/Japan Agency for Medical Research and Development (AMED)
- JP20H00629/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP23H00516/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP18H03372/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Miscellaneous

