Transcription-coupled DNA-protein crosslink repair by CSB and CRL4CSA-mediated degradation
- PMID: 38600236
- PMCID: PMC11098752
- DOI: 10.1038/s41556-024-01394-y
Transcription-coupled DNA-protein crosslink repair by CSB and CRL4CSA-mediated degradation
Abstract
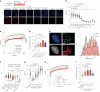
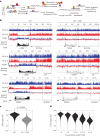
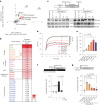
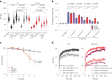
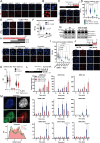
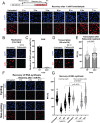
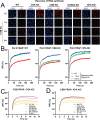
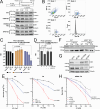
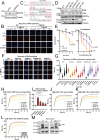
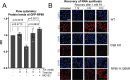
DNA-protein crosslinks (DPCs) arise from enzymatic intermediates, metabolism or chemicals like chemotherapeutics. DPCs are highly cytotoxic as they impede DNA-based processes such as replication, which is counteracted through proteolysis-mediated DPC removal by spartan (SPRTN) or the proteasome. However, whether DPCs affect transcription and how transcription-blocking DPCs are repaired remains largely unknown. Here we show that DPCs severely impede RNA polymerase II-mediated transcription and are preferentially repaired in active genes by transcription-coupled DPC (TC-DPC) repair. TC-DPC repair is initiated by recruiting the transcription-coupled nucleotide excision repair (TC-NER) factors CSB and CSA to DPC-stalled RNA polymerase II. CSA and CSB are indispensable for TC-DPC repair; however, the downstream TC-NER factors UVSSA and XPA are not, a result indicative of a non-canonical TC-NER mechanism. TC-DPC repair functions independently of SPRTN but is mediated by the ubiquitin ligase CRL4CSA and the proteasome. Thus, DPCs in genes are preferentially repaired in a transcription-coupled manner to facilitate unperturbed transcription.
© 2024. The Author(s).
Conflict of interest statement
S.C.D. is President/CEO of DeaTech Research, an engineering and biotechnology consulting firm, but has no financial or intellectual property interests in this study. The other authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

