Cell-type-resolved mosaicism reveals clonal dynamics of the human forebrain
- PMID: 38600385
- PMCID: PMC11194162
- DOI: 10.1038/s41586-024-07292-5
Cell-type-resolved mosaicism reveals clonal dynamics of the human forebrain
Abstract
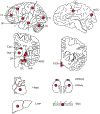
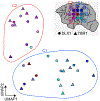
Debate remains around the anatomical origins of specific brain cell subtypes and lineage relationships within the human forebrain1-7. Thus, direct observation in the mature human brain is critical for a complete understanding of its structural organization and cellular origins. Here we utilize brain mosaic variation within specific cell types as distinct indicators for clonal dynamics, denoted as cell-type-specific mosaic variant barcode analysis. From four hemispheres and two different human neurotypical donors, we identified 287 and 780 mosaic variants, respectively, that were used to deconvolve clonal dynamics. Clonal spread and allele fractions within the brain reveal that local hippocampal excitatory neurons are more lineage-restricted than resident neocortical excitatory neurons or resident basal ganglia GABAergic inhibitory neurons. Furthermore, simultaneous genome transcriptome analysis at both a cell-type-specific and a single-cell level suggests a dorsal neocortical origin for a subgroup of DLX1+ inhibitory neurons that disperse radially from an origin shared with excitatory neurons. Finally, the distribution of mosaic variants across 17 locations within one parietal lobe reveals that restriction of clonal spread in the anterior-posterior axis precedes restriction in the dorsal-ventral axis for both excitatory and inhibitory neurons. Thus, cell-type-resolved somatic mosaicism can uncover lineage relationships governing the development of the human forebrain.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
K.K. is a senior scientist at Bioskryb Genomics Inc. All other authors declare no competing interests.
Figures













Update of
-
Cell-type-resolved somatic mosaicism reveals clonal dynamics of the human forebrain.bioRxiv [Preprint]. 2023 Oct 26:2023.10.24.563814. doi: 10.1101/2023.10.24.563814. bioRxiv. 2023. Update in: Nature. 2024 May;629(8011):384-392. doi: 10.1038/s41586-024-07292-5. PMID: 37961480 Free PMC article. Updated. Preprint.

