Long noncoding RNAs and mRNAs profiling in ovary during laying and broodiness in Taihe Black-Bone Silky Fowls (Gallus gallus Domesticus Brisson)
- PMID: 38600449
- PMCID: PMC11005167
- DOI: 10.1186/s12864-024-10281-7
Long noncoding RNAs and mRNAs profiling in ovary during laying and broodiness in Taihe Black-Bone Silky Fowls (Gallus gallus Domesticus Brisson)
Abstract
Background: Broodiness significantly impacts poultry egg production, particularly notable in specific breeds such as the black-boned Silky, characterized by pronounced broodiness. An understanding of the alterations in ovarian signaling is essential for elucidating the mechanisms that influence broodiness. However, comparative research on the characteristics of long non-coding RNAs (lncRNAs) in the ovaries of broody chickens (BC) and high egg-laying chickens (GC) remains scant. In this investigation, we employed RNA sequencing to assess the ovarian transcriptomes, which include both lncRNAs and mRNAs, in eight Taihe Black-Bone Silky Fowls (TBsf), categorized into broody and high egg-laying groups. This study aims to provide a clearer understanding of the genetic underpinnings associated with broodiness and egg production.
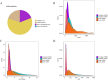
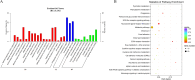
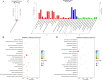
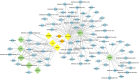
Results: We have identified a total of 16,444 mRNAs and 18,756 lncRNAs, of which 349 mRNAs and 651 lncRNAs exhibited significantly different expression (DE) between the BC and GC groups. Furthermore, we have identified the cis-regulated and trans-regulated target genes of differentially abundant lncRNA transcripts and have constructed an lncRNA-mRNA trans-regulated interaction network linked to ovarian follicle development. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation analyses have revealed that DE mRNAs and the target genes of DE lncRNAs are associated with pathways including neuroactive ligand-receptor interaction, CCR6 chemokine receptor binding, G-protein coupled receptor binding, cytokine-cytokine receptor interaction, and ECM-receptor interaction.
Conclusion: Our research presents a comprehensive compilation of lncRNAs and mRNAs linked to ovarian development. Additionally, it establishes a predictive interaction network involving differentially abundant lncRNAs and differentially expressed genes (DEGs) within TBsf. This significantly contributes to our understanding of the intricate interactions between lncRNAs and genes governing brooding behavior.
Keywords: Broodiness; Egg production; Ovarian follicle development; Transcriptomes; lncRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Long noncoding RNAs profiling in ovary during laying and nesting in Muscovy ducks (Cairina moschata).Anim Reprod Sci. 2021 Jul;230:106762. doi: 10.1016/j.anireprosci.2021.106762. Epub 2021 May 8. Anim Reprod Sci. 2021. PMID: 34022609
-
Proteomic analysis of egg production peak and senescence in the ovaries of Taihe black-boned silky fowl (Gallus gallus domesticus Brisson).BMC Genomics. 2025 Jan 7;26(1):17. doi: 10.1186/s12864-024-11180-7. BMC Genomics. 2025. PMID: 39773120 Free PMC article.
-
Identification and functional analysis of ovarian lncRNAs during different egg laying periods in Taihe Black-Bone Chickens.Front Physiol. 2024 Feb 15;15:1358682. doi: 10.3389/fphys.2024.1358682. eCollection 2024. Front Physiol. 2024. PMID: 38426211 Free PMC article.
-
Transcriptome Analysis of the Ovaries of Taihe Black-Bone Silky Fowls at Different Egg-Laying Stages.Genes (Basel). 2022 Nov 8;13(11):2066. doi: 10.3390/genes13112066. Genes (Basel). 2022. PMID: 36360303 Free PMC article.
-
Integrating Genomics and Transcriptomics to Identify Candidate Genes for Egg Production in Taihe Black-Bone Silky Fowls (Gallus gallus domesticus Brisson).Int J Mol Sci. 2024 Aug 29;25(17):9373. doi: 10.3390/ijms25179373. Int J Mol Sci. 2024. PMID: 39273321 Free PMC article.
Cited by
-
Frontiers and emerging topics in a century of Silkie chicken research: insights, challenges, and opportunities.Poult Sci. 2025 May;104(5):105030. doi: 10.1016/j.psj.2025.105030. Epub 2025 Mar 12. Poult Sci. 2025. PMID: 40101517 Free PMC article. Review.
-
Lnc MSTRG 4701.7 targets miR-1786/RORa to competitively regulate proliferation and apoptosis in chicken follicular granulosa cells.Front Vet Sci. 2025 Apr 30;12:1583287. doi: 10.3389/fvets.2025.1583287. eCollection 2025. Front Vet Sci. 2025. PMID: 40370832 Free PMC article.
-
Ovarian Transcriptome Profile from Egg-Laying Period to Incubation Period of Changshun Green-Shell Laying Hens.Genes (Basel). 2025 Mar 29;16(4):394. doi: 10.3390/genes16040394. Genes (Basel). 2025. PMID: 40282353 Free PMC article.
References
-
- Zhang L, Cui X. Nesting behavior of black chicken and artificial forced molting technology. Jilin xu mu shou yi. 2018;39:32+34.
-
- Mn R, Rt T, Pw W, Pj S. Genetic control of incubation behavior in the domestic hen. Poult Sci. 2002;81 10.1093/ps/81.7.928 - PubMed
MeSH terms
Substances
Grants and funding
- 2021-KYY-517102-0023/Major Scientific and Technological cooperation between Zhejiang University and Taihe County Government
- 2021-KYY-517102-0023/Major Scientific and Technological cooperation between Zhejiang University and Taihe County Government
- 2021-KYY-517102-0023/Major Scientific and Technological cooperation between Zhejiang University and Taihe County Government
- 2021-KYY-517102-0023/Major Scientific and Technological cooperation between Zhejiang University and Taihe County Government
- 2021-KYY-517102-0023/Major Scientific and Technological cooperation between Zhejiang University and Taihe County Government
- 2021-KYY-517102-0023/Major Scientific and Technological cooperation between Zhejiang University and Taihe County Government
- 2021C02068-11/the Zhejiang Provincial "Fourteenth Five Year Plan" major scientific and technological special projects in agriculture
- 2021C02068-11/the Zhejiang Provincial "Fourteenth Five Year Plan" major scientific and technological special projects in agriculture
- 2021C02068-11/the Zhejiang Provincial "Fourteenth Five Year Plan" major scientific and technological special projects in agriculture
- 2021C02068-11/the Zhejiang Provincial "Fourteenth Five Year Plan" major scientific and technological special projects in agriculture
- 2021C02068-11/the Zhejiang Provincial "Fourteenth Five Year Plan" major scientific and technological special projects in agriculture
- 2021C02068-11/the Zhejiang Provincial "Fourteenth Five Year Plan" major scientific and technological special projects in agriculture
LinkOut - more resources
Full Text Sources
Miscellaneous

