Clinical manifestations, antimicrobial resistance and genomic feature analysis of multidrug-resistant Elizabethkingia strains
- PMID: 38600542
- PMCID: PMC11007976
- DOI: 10.1186/s12941-024-00691-6
Clinical manifestations, antimicrobial resistance and genomic feature analysis of multidrug-resistant Elizabethkingia strains
Abstract
Background: Elizabethkingia is emerging as an opportunistic pathogen in humans. The aim of this study was to investigate the clinical epidemiology, antimicrobial susceptibility, virulence factors, and genome features of Elizabethkingia spp.
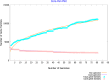
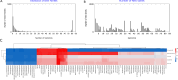
Methods: Clinical data from 71 patients who were diagnosed with Elizabethkingia-induced pneumonia and bacteremia between August 2019 and September 2021 were analyzed. Whole-genome sequencing was performed on seven isolates, and the results were compared with a dataset of 83 available Elizabethkingia genomes. Genomic features, Kyoto Encyclopedia of Genes and Genomes (KEGG) results and clusters of orthologous groups (COGs) were analyzed.
Results: The mean age of the patients was 56.9 ± 20.7 years, and the in-hospital mortality rate was 29.6% (21/71). Elizabethkingia strains were obtained mainly from intensive care units (36.6%, 26/71) and emergency departments (32.4%, 23/71). The majority of the strains were isolated from respiratory tract specimens (85.9%, 61/71). All patients had a history of broad-spectrum antimicrobial exposure. Hospitalization for invasive mechanical ventilation or catheter insertion was found to be a risk factor for infection. The isolates displayed a high rate of resistance to cephalosporins and carbapenems, but all were susceptible to minocycline and colistin. Genomic analysis identified five β-lactamase genes (blaGOB, blaBlaB, blaCME, blaOXA, and blaTEM) responsible for β-lactam resistance and virulence genes involved in stress adaptation (ureB/G, katA/B, and clpP), adherence (groEL, tufA, and htpB) and immune modulation (gmd, tviB, cps4J, wbtIL, cap8E/D/G, and rfbC). Functional analysis of the COGs revealed that "metabolism" constituted the largest category within the core genome, while "information storage and processing" was predominant in both the accessory and unique genomes. The unique genes in our 7 strains were mostly enriched in KEGG pathways related to microRNAs in cancer, drug resistance (β-lactam and vancomycin), ABC transporters, biological metabolism and biosynthesis, and nucleotide excision repair mechanisms.
Conclusion: The Elizabethkingia genus exhibits multidrug resistance and carries carbapenemase genes. This study presents a comparative genomic analysis of Elizabethkingia, providing knowledge that facilitates a better understanding of this microorganism.
Keywords: Elizabethkingia; Antimicrobial susceptibility; Carbapenem resistance; Genomic analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

References
-
- Kim KK, Kim MK, Lim JH, Park HY, Lee ST. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. Nov. Int J Syst Evol MicroBiol. 2005;55(Pt 3):1287–93. doi: 10.1099/ijs.0.63541-0. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

