Targeting FTO induces colorectal cancer ferroptotic cell death by decreasing SLC7A11/GPX4 expression
- PMID: 38600610
- PMCID: PMC11005233
- DOI: 10.1186/s13046-024-03032-9
Targeting FTO induces colorectal cancer ferroptotic cell death by decreasing SLC7A11/GPX4 expression
Erratum in
-
Correction: Targeting FTO induces colorectal cancer ferroptotic cell death by decreasing SLC7A11/ GPX4 expression.J Exp Clin Cancer Res. 2024 May 2;43(1):131. doi: 10.1186/s13046-024-03051-6. J Exp Clin Cancer Res. 2024. PMID: 38693552 Free PMC article. No abstract available.
Abstract
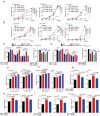
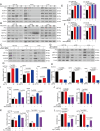
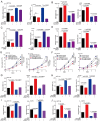
Ferroptosis is a newly identified iron-dependent form of death that is becoming increasingly recognized as a promising avenue for cancer therapy. N6-methyladenosine (m6A) is the most abundant reversible methylation modification in mRNA contributing to tumorigenesis. However, the crucial role of m6A modification in regulating ferroptosis during colorectal cancer (CRC) tumorigenesis remains elusive. Herein, we find that m6A modification is increased during ferroptotic cell death and correlates with the decreased m6A demethylase fat mass and obesity-associated protein (FTO) expression. Functionally, we demonstrate that suppressing FTO significantly induces CRC ferroptotic cell death, as well as enhancing CRC cell sensitivity to ferroptosis inducer (Erastin and RSL3) treatment. Mechanistically, high FTO expression increased solute carrier family 7 member 11 (SLC7A11) or glutathione peroxidase 4 (GPX4) expressions in an m6A-YTHDF2 dependent manner, thereby counteracting ferroptotic cell death stress. In addition, we identify Mupirocin as a novel inhibitor of FTO, and Mupirocin induces CRC ferroptosis and inhibits tumor growth. Clinically, the levels of FTO, SLC7A11, and GPX4, are highly correlated expression in CRC tissues. Our findings reveal that FTO protects CRC from ferroptotic cell death in promoting CRC tumorigenesis through triggering SLC7A11/GPX4 expression.
Keywords: Colorectal cancer (CRC); Fat mass and obesity-associated protein (FTO); Ferroptosis; Glutathione peroxidase 4 (GPX4); N6-methyladenosine (m6A); Solute carrier family 7 member 11 (SLC7A11).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures




References
-
- Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409–436. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

