Bioinformatics and system biology approach to identify the influences among COVID-19, influenza, and HIV on the regulation of gene expression
- PMID: 38601162
- PMCID: PMC11004287
- DOI: 10.3389/fimmu.2024.1369311
Bioinformatics and system biology approach to identify the influences among COVID-19, influenza, and HIV on the regulation of gene expression
Abstract
Background: Coronavirus disease (COVID-19), caused by SARS-CoV-2, has emerged as a infectious disease, coexisting with widespread seasonal and sporadic influenza epidemics globally. Individuals living with HIV, characterized by compromised immune systems, face an elevated risk of severe outcomes and increased mortality when affected by COVID-19. Despite this connection, the molecular intricacies linking COVID-19, influenza, and HIV remain unclear. Our research endeavors to elucidate the shared pathways and molecular markers in individuals with HIV concurrently infected with COVID-19 and influenza. Furthermore, we aim to identify potential medications that may prove beneficial in managing these three interconnected illnesses.
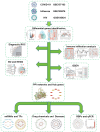
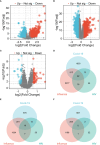
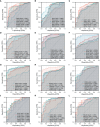
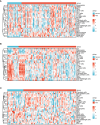
Methods: Sequencing data for COVID-19 (GSE157103), influenza (GSE185576), and HIV (GSE195434) were retrieved from the GEO database. Commonly expressed differentially expressed genes (DEGs) were identified across the three datasets, followed by immune infiltration analysis and diagnostic ROC analysis on the DEGs. Functional enrichment analysis was performed using GO/KEGG and Gene Set Enrichment Analysis (GSEA). Hub genes were screened through a Protein-Protein Interaction networks (PPIs) analysis among DEGs. Analysis of miRNAs, transcription factors, drug chemicals, diseases, and RNA-binding proteins was conducted based on the identified hub genes. Finally, quantitative PCR (qPCR) expression verification was undertaken for selected hub genes.
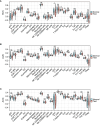
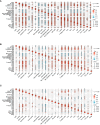
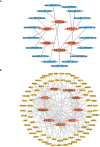
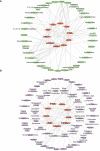
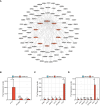
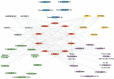
Results: The analysis of the three datasets revealed a total of 22 shared DEGs, with the majority exhibiting an area under the curve value exceeding 0.7. Functional enrichment analysis with GO/KEGG and GSEA primarily highlighted signaling pathways associated with ribosomes and tumors. The ten identified hub genes included IFI44L, IFI44, RSAD2, ISG15, IFIT3, OAS1, EIF2AK2, IFI27, OASL, and EPSTI1. Additionally, five crucial miRNAs (hsa-miR-8060, hsa-miR-6890-5p, hsa-miR-5003-3p, hsa-miR-6893-3p, and hsa-miR-6069), five essential transcription factors (CREB1, CEBPB, EGR1, EP300, and IRF1), and the top ten significant drug chemicals (estradiol, progesterone, tretinoin, calcitriol, fluorouracil, methotrexate, lipopolysaccharide, valproic acid, silicon dioxide, cyclosporine) were identified.
Conclusion: This research provides valuable insights into shared molecular targets, signaling pathways, drug chemicals, and potential biomarkers for individuals facing the complex intersection of COVID-19, influenza, and HIV. These findings hold promise for enhancing the precision of diagnosis and treatment for individuals with HIV co-infected with COVID-19 and influenza.
Keywords: COVID-19; HIV; differentially expressed genes; drug chemicals; hub genes; immune infiltration; influenza; protein-protein interaction networks.
Copyright © 2024 Zhang, Jin, Zhang, Bai, Zheng, Tian, Deng, Mao, Qiu and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

