An Epigenome-Wide Association Study of DNA Methylation and Proliferative Retinopathy over 28 Years in Type 1 Diabetes
- PMID: 38601260
- PMCID: PMC11004204
- DOI: 10.1016/j.xops.2024.100497
An Epigenome-Wide Association Study of DNA Methylation and Proliferative Retinopathy over 28 Years in Type 1 Diabetes
Abstract
Purpose: To perform a prospective epigenome-wide association study of DNA methylation (DNAm) and 28-year proliferative diabetic retinopathy (PDR) incidence in type 1 diabetes (T1D).
Design: Prospective observational cohort study.
Participants: The Pittsburgh Epidemiology of Diabetes Complications (EDC) study of childhood-onset (< 17 years) T1D.
Methods: Stereoscopic fundus photographs were taken in fields 1, 2, and 4 at baseline, 2, 4, 6, 8, 16, 23, and 28 years after DNAm measurements. The photos were graded using the modified Airlie House System. In those free of PDR at baseline (n = 265; mean T1D duration of 18 years at baseline), whole blood DNAm (EPIC array) at 683 597 CpGs was analyzed in Cox models for time to event. Associations between significant CpGs and clinical risk factors were assessed; genetic variants associated with DNAm were identified (methylation quantitative trait loci [meQTLs]). Mendelian randomization was used to examine evidence of causal associations between DNAm and PDR. Post hoc regional and functional analyses were performed.
Main outcome measures: Proliferative diabetic retinopathy was defined as the first instance of a grade of ≥ 60 in at least 1 eye or pan-retinal photocoagulation for PDR. Follow-up time was calculated from the study visit at which DNAm data were available (baseline) until PDR incidence or censoring (December 31, 2018 or last follow-up).
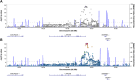
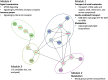
Results: PDR incidence was 53% over 28-years' follow-up. Greater DNAm of cg27512687 (KIF16B) was associated with reduced PDR incidence (P = 6.3 × 10-9; false discovery rate [FDR]: < 0.01); 113 cis-meQTLs (P < 5 × 10-8) were identified. Mendelian randomization analysis using the sentinel meQTL as the instrumental variable supported a potentially causal association between cg27512687 and PDR. Cg27512687 was also associated with lower pulse rate and albumin excretion rate and higher estimated glomerular filtration rate, but its association with PDR remained independently significant after adjustment for those factors. In regional analyses, DNAm of FUT4, FKBP1A, and RIN2 was also associated with PDR incidence.
Conclusions: DNA methylation of KIF16B, FUT4, FKBP1A, and RIN2 was associated with PDR incidence, supporting roles for epigenetic regulation of iron clearance, developmental pathways, and autophagy in PDR pathogenesis. Further study of those loci may provide insight into novel targets for interventions to prevent or delay PDR in T1D.
Financial disclosures: Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: DNA methylation; Epigenetics; Proliferative diabetic retinopathy; Retina; Type 1 diabetes.
© 2024 by the American Academy of Ophthalmology.
Figures
Similar articles
-
DNA methylation and 28-year cardiovascular disease risk in type 1 diabetes: the Epidemiology of Diabetes Complications (EDC) cohort study.Clin Epigenetics. 2023 Aug 2;15(1):122. doi: 10.1186/s13148-023-01539-0. Clin Epigenetics. 2023. PMID: 37533055 Free PMC article.
-
Prognostic factors for the development and progression of proliferative diabetic retinopathy in people with diabetic retinopathy.Cochrane Database Syst Rev. 2023 Feb 22;2(2):CD013775. doi: 10.1002/14651858.CD013775.pub2. Cochrane Database Syst Rev. 2023. PMID: 36815723 Free PMC article. Review.
-
Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy.BMC Med. 2015 Aug 6;13:182. doi: 10.1186/s12916-015-0421-5. BMC Med. 2015. PMID: 26248552 Free PMC article.
-
DNA Methylation and 28-Year Incidence of Kidney Disease in Type 1 Diabetes.Kidney360. 2025 Jun 9. doi: 10.34067/KID.0000000881. Online ahead of print. Kidney360. 2025. PMID: 40489246
-
Proliferative retinopathy during hyperbaric oxygen treatment.Diving Hyperb Med. 2017 Sep;47(3):203. doi: 10.28920/dhm47.3.203. Diving Hyperb Med. 2017. PMID: 28868603 Free PMC article. Review.
Cited by
-
DNA methylation and 28 year incidence of two neuropathy phenotypes in type 1 diabetes: the Pittsburgh Epidemiology of Diabetes Complications cohort study.Diabetologia. 2025 Jul;68(7):1423-1439. doi: 10.1007/s00125-025-06427-1. Epub 2025 Apr 23. Diabetologia. 2025. PMID: 40266295
-
Epigenetics Plays a Role in the Pathogenesis and Treatment of Diabetes.Genes (Basel). 2025 Jun 30;16(7):769. doi: 10.3390/genes16070769. Genes (Basel). 2025. PMID: 40725425 Free PMC article. Review.
References
-
- Zhang L., Krzentowski G., Albert A., Lefebvre P.J. Risk of developing retinopathy in diabetes control and complications trial type 1 diabetic patients with good or poor metabolic control. Diabetes Care. 2001;24:1275–1279. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

