The role of RASA2 in predicting radioresistance in lung cancer through regulation of p53
- PMID: 38601440
- PMCID: PMC11002505
- DOI: 10.21037/tlcr-24-160
The role of RASA2 in predicting radioresistance in lung cancer through regulation of p53
Abstract
Background: One of the most common causes of lung cancer relapse after clinical treatment is radioresistance. However, the mechanism underlying radioresistance remains unclear. In this study, we investigated the role of Ras p21 protein activator (RASA2) in non-small cell lung cancer (NSCLC).
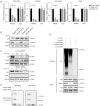
Methods: The messenger RNA (mRNA) of RASA2 was tested via reverse-transcription quantitative polymerase chain reaction (RT-qPCR) of cancer tissues from patients with NSCLC. Computed tomography (CT) and bioluminescent imaging (BLI) were used to monitor the tumor growth of patients and orthotopic mice, respectively. Protein-protein interaction was quantified via immunoprecipitation and glutathione S transferase (GST) pulldown assay. Western blotting was used to evaluate the phosphorylation and ubiquitination level of p53.
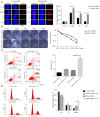
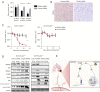
Results: The results indicated a negative correlation between the mRNA expression levels of RASA2 in tumor tissues with patients' response to radiotherapy. Patients with a high expression of RASA2 had a lower objective response rate (ORR) after 1 month of radiotherapy than patients with low expression of RASA2 after 1 month of radiotherapy. In terms of mechanism, we proved that RASA2 can directly bind to p53 to promote the phosphorylation of p53, which inhibits its transcriptional activity and further promotes its degradation through the ubiquitin/proteasome pathway. In this process, the apoptosis of tumor cells is inhibited due to impaired p53 surveillance, which leads to radioresistance.
Conclusions: Our results demonstrate that RASA2 negatively regulates p53 in cancer cells and therefore promotes radioresistance, providing a new predictive biomarker and a potential therapeutic target for radioresistance.
Keywords: Ras p21 protein activator (RASA2); non-small cell lung cancer (NSCLC); p53; radioresistance.
2024 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-160/coif). The authors have no conflicts of interest to declare.
Figures



Similar articles
-
CDK16 Phosphorylates and Degrades p53 to Promote Radioresistance and Predicts Prognosis in Lung Cancer.Theranostics. 2018 Jan 1;8(3):650-662. doi: 10.7150/thno.21963. eCollection 2018. Theranostics. 2018. PMID: 29344296 Free PMC article.
-
SDH5 Depletion Enhances Radiosensitivity by Regulating p53: A New Method for Noninvasive Prediction of Radiotherapy Response.Theranostics. 2019 Aug 14;9(22):6380-6395. doi: 10.7150/thno.34443. eCollection 2019. Theranostics. 2019. PMID: 31588224 Free PMC article.
-
The kinesin light chain-2, a target of mRNA stabilizing protein HuR, inhibits p53 protein phosphorylation to promote radioresistance in NSCLC.Thorac Cancer. 2023 Jun;14(16):1440-1450. doi: 10.1111/1759-7714.14886. Epub 2023 Apr 13. Thorac Cancer. 2023. PMID: 37055376 Free PMC article.
-
CLPTM1L induces estrogen receptor β signaling-mediated radioresistance in non-small cell lung cancer cells.Cell Commun Signal. 2020 Sep 17;18(1):152. doi: 10.1186/s12964-020-00571-4. Cell Commun Signal. 2020. PMID: 32943060 Free PMC article.
-
P53-mediated radioresistance does not correlate with metastatic potential in tumorigenic rat embryo cell lines following oncogene transfection.Int J Radiat Oncol Biol Phys. 1996 Jan 15;34(2):341-55. doi: 10.1016/0360-3016(95)02023-3. Int J Radiat Oncol Biol Phys. 1996. PMID: 8567335 Review.
Cited by
-
Unveiling the impact of glutathione (GSH) and p53 gene deletion on tumor cell metabolism by amino acid and proteomics analysis.J Gastrointest Oncol. 2024 Jun 30;15(3):1002-1019. doi: 10.21037/jgo-24-236. Epub 2024 Jun 27. J Gastrointest Oncol. 2024. PMID: 38989407 Free PMC article.
-
Re-irradiation for local recurrence after definitive stereotactic body radiotherapy for early-stage non-small cell lung cancer.Transl Lung Cancer Res. 2025 May 30;14(5):1650-1659. doi: 10.21037/tlcr-2025-89. Epub 2025 May 27. Transl Lung Cancer Res. 2025. PMID: 40535073 Free PMC article.
-
Nuclear receptors as novel regulators that modulate cancer radiosensitivity and normal tissue radiotoxicity.Mol Cancer. 2025 May 30;24(1):155. doi: 10.1186/s12943-025-02362-2. Mol Cancer. 2025. PMID: 40442680 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
