Deep learning or radiomics based on CT for predicting the response of gastric cancer to neoadjuvant chemotherapy: a meta-analysis and systematic review
- PMID: 38601765
- PMCID: PMC11004479
- DOI: 10.3389/fonc.2024.1363812
Deep learning or radiomics based on CT for predicting the response of gastric cancer to neoadjuvant chemotherapy: a meta-analysis and systematic review
Erratum in
-
Corrigendum: Deep learning or radiomics based on CT for predicting the response of gastric cancer to neoadjuvant chemotherapy: a meta-analysis and systematic review.Front Oncol. 2024 May 23;14:1433346. doi: 10.3389/fonc.2024.1433346. eCollection 2024. Front Oncol. 2024. PMID: 38846979 Free PMC article.
Abstract
Background: Artificial intelligence (AI) models, clinical models (CM), and the integrated model (IM) are utilized to evaluate the response to neoadjuvant chemotherapy (NACT) in patients diagnosed with gastric cancer.
Objective: The objective is to identify the diagnostic test of the AI model and to compare the accuracy of AI, CM, and IM through a comprehensive summary of head-to-head comparative studies.
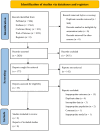
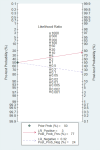
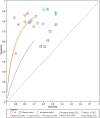
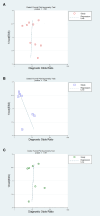
Methods: PubMed, Web of Science, Cochrane Library, and Embase were systematically searched until September 5, 2023, to compile English language studies without regional restrictions. The quality of the included studies was evaluated using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) criteria. Forest plots were utilized to illustrate the findings of diagnostic accuracy, while Hierarchical Summary Receiver Operating Characteristic curves were generated to estimate sensitivity (SEN) and specificity (SPE). Meta-regression was applied to analyze heterogeneity across the studies. To assess the presence of publication bias, Deeks' funnel plot and an asymmetry test were employed.
Results: A total of 9 studies, comprising 3313 patients, were included for the AI model, with 7 head-to-head comparative studies involving 2699 patients. Across the 9 studies, the pooled SEN for the AI model was 0.75 (95% confidence interval (CI): 0.66, 0.82), and SPE was 0.77 (95% CI: 0.69, 0.84). Meta-regression was conducted, revealing that the cut-off value, approach to predicting response, and gold standard might be sources of heterogeneity. In the head-to-head comparative studies, the pooled SEN for AI was 0.77 (95% CI: 0.69, 0.84) with SPE at 0.79 (95% CI: 0.70, 0.85). For CM, the pooled SEN was 0.67 (95% CI: 0.57, 0.77) with SPE at 0.59 (95% CI: 0.54, 0.64), while for IM, the pooled SEN was 0.83 (95% CI: 0.79, 0.86) with SPE at 0.69 (95% CI: 0.56, 0.79). Notably, there was no statistical difference, except that IM exhibited higher SEN than AI, while maintaining a similar level of SPE in pairwise comparisons. In the Receiver Operating Characteristic analysis subgroup, the CT-based Deep Learning (DL) subgroup, and the National Comprehensive Cancer Network (NCCN) guideline subgroup, the AI model exhibited higher SEN but lower SPE compared to the IM. Conversely, in the training cohort subgroup and the internal validation cohort subgroup, the AI model demonstrated lower SEN but higher SPE than the IM. The subgroup analysis underscored that factors such as the number of cohorts, cohort type, cut-off value, approach to predicting response, and choice of gold standard could impact the reliability and robustness of the results.
Conclusion: AI has demonstrated its viability as a tool for predicting the response of GC patients to NACT Furthermore, CT-based DL model in AI was sensitive to extract tumor features and predict the response. The results of subgroup analysis also supported the above conclusions. Large-scale rigorously designed diagnostic accuracy studies and head-to-head comparative studies are anticipated.
Systematic review registration: PROSPERO, CRD42022377030.
Keywords: artificial intelligence; deep learning; gastric cancer; meta-analysis; neoadjuvant chemotherapy; radiomics.
Copyright © 2024 Bao, Du, Zheng, Guo and Ji.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Convolutional neural network-based artificial intelligence for the diagnosis of early esophageal cancer based on endoscopic images: A meta-analysis.Saudi J Gastroenterol. 2022 Sep-Oct;28(5):332-340. doi: 10.4103/sjg.sjg_178_22. Saudi J Gastroenterol. 2022. PMID: 35848703 Free PMC article. Review.
-
Artificial intelligence with magnetic resonance imaging for prediction of pathological complete response to neoadjuvant chemoradiotherapy in rectal cancer: A systematic review and meta-analysis.Front Oncol. 2022 Oct 12;12:1026216. doi: 10.3389/fonc.2022.1026216. eCollection 2022. Front Oncol. 2022. PMID: 36313696 Free PMC article.
-
MRI-based radiomics for predicting pathological complete response after neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a systematic review and meta-analysis.Front Oncol. 2025 Mar 10;15:1550838. doi: 10.3389/fonc.2025.1550838. eCollection 2025. Front Oncol. 2025. PMID: 40129922 Free PMC article.
-
Using artificial intelligence based imaging to predict lymph node metastasis in non-small cell lung cancer: a systematic review and meta-analysis.Quant Imaging Med Surg. 2024 Oct 1;14(10):7496-7512. doi: 10.21037/qims-24-664. Epub 2024 Sep 26. Quant Imaging Med Surg. 2024. PMID: 39429617 Free PMC article.
-
[Diagnostic value of optical imaging combined with indocyanine green-guided sentinel lymph node biopsy in gastric cancer: a meta-analysis].Zhonghua Wei Chang Wai Ke Za Zhi. 2019 Dec 25;22(12):1196-1204. doi: 10.3760/cma.j.issn.1671-0274.2019.12.017. Zhonghua Wei Chang Wai Ke Za Zhi. 2019. PMID: 31874538 Chinese.
Cited by
-
Artificial intelligence as a predictive tool for gastric cancer: Bridging innovation, clinical translation, and ethical considerations.World J Gastrointest Oncol. 2025 May 15;17(5):103275. doi: 10.4251/wjgo.v17.i5.103275. World J Gastrointest Oncol. 2025. PMID: 40487933 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

