Coronary Artery Disease Risk Variant Dampens the Expression of CALCRL by Reducing HSF Binding to Shear Stress Responsive Enhancer in Endothelial Cells In Vitro
- PMID: 38602103
- PMCID: PMC11111333
- DOI: 10.1161/ATVBAHA.123.318964
Coronary Artery Disease Risk Variant Dampens the Expression of CALCRL by Reducing HSF Binding to Shear Stress Responsive Enhancer in Endothelial Cells In Vitro
Abstract
Background: CALCRL (calcitonin receptor-like) protein is an important mediator of the endothelial fluid shear stress response, which is associated with the genetic risk of coronary artery disease. In this study, we functionally characterized the noncoding regulatory elements carrying coronary artery disease that risks single-nucleotide polymorphisms and studied their role in the regulation of CALCRL expression in endothelial cells.
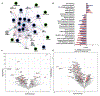
Methods: To functionally characterize the coronary artery disease single-nucleotide polymorphisms harbored around the gene CALCRL, we applied an integrative approach encompassing statistical, transcriptional (RNA-seq), and epigenetic (ATAC-seq [transposase-accessible chromatin with sequencing], chromatin immunoprecipitation assay-quantitative polymerase chain reaction, and electromobility shift assay) analyses, alongside luciferase reporter assays, and targeted gene and enhancer perturbations (siRNA and clustered regularly interspaced short palindromic repeats/clustered regularly interspaced short palindromic repeat-associated 9) in human aortic endothelial cells.
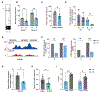
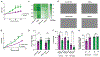
Results: We demonstrate that the regulatory element harboring rs880890 exhibits high enhancer activity and shows significant allelic bias. The A allele was favored over the G allele, particularly under shear stress conditions, mediated through alterations in the HSF1 (heat shock factor 1) motif and binding. CRISPR deletion of rs880890 enhancer resulted in downregulation of CALCRL expression, whereas HSF1 knockdown resulted in a significant decrease in rs880890-enhancer activity and CALCRL expression. A significant decrease in HSF1 binding to the enhancer region in endothelial cells was observed under disturbed flow compared with unidirectional flow. CALCRL knockdown and variant perturbation experiments indicated the role of CALCRL in mediating eNOS (endothelial nitric oxide synthase), APLN (apelin), angiopoietin, prostaglandins, and EDN1 (endothelin-1) signaling pathways leading to a decrease in cell proliferation, tube formation, and NO production.
Conclusions: Overall, our results demonstrate the existence of an endothelial-specific HSF (heat shock factor)-regulated transcriptional enhancer that mediates CALCRL expression. A better understanding of CALCRL gene regulation and the role of single-nucleotide polymorphisms in the modulation of CALCRL expression could provide important steps toward understanding the genetic regulation of shear stress signaling responses.
Keywords: coronary artery disease; coronary vessels; gene expression; genome-wide association study; polymorphism, single nucleotide.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

