Tumor Biomechanics Alters Metastatic Dissemination of Triple Negative Breast Cancer via Rewiring Fatty Acid Metabolism
- PMID: 38602451
- PMCID: PMC11186052
- DOI: 10.1002/advs.202307963
Tumor Biomechanics Alters Metastatic Dissemination of Triple Negative Breast Cancer via Rewiring Fatty Acid Metabolism
Abstract
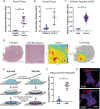
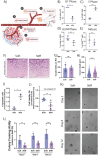
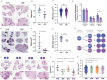
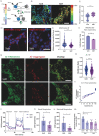
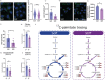
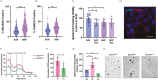
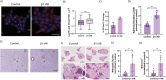
In recent decades, the role of tumor biomechanics on cancer cell behavior at the primary site has been increasingly appreciated. However, the effect of primary tumor biomechanics on the latter stages of the metastatic cascade, such as metastatic seeding of secondary sites and outgrowth remains underappreciated. This work sought to address this in the context of triple negative breast cancer (TNBC), a cancer type known to aggressively disseminate at all stages of disease progression. Using mechanically tuneable model systems, mimicking the range of stiffness's typically found within breast tumors, it is found that, contrary to expectations, cancer cells exposed to softer microenvironments are more able to colonize secondary tissues. It is shown that heightened cell survival is driven by enhanced metabolism of fatty acids within TNBC cells exposed to softer microenvironments. It is demonstrated that uncoupling cellular mechanosensing through integrin β1 blocking antibody effectively causes stiff primed TNBC cells to behave like their soft counterparts, both in vitro and in vivo. This work is the first to show that softer tumor microenvironments may be contributing to changes in disease outcome by imprinting on TNBC cells a greater metabolic flexibility and conferring discrete cell survival advantages.
Keywords: biomechanics; breast cancer; extracellular matrix; metabolism; metastasis.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Cox T. R., Nat. Rev. Cancer 2021, 21, 217. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
