Hox gene-specific cellular targeting using split intein Trojan exons
- PMID: 38602904
- PMCID: PMC11047080
- DOI: 10.1073/pnas.2317083121
Hox gene-specific cellular targeting using split intein Trojan exons
Abstract
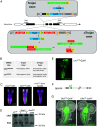
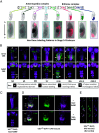
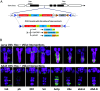
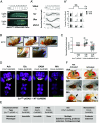
The Trojan exon method, which makes use of intronically inserted T2A-Gal4 cassettes, has been widely used in Drosophila to create thousands of gene-specific Gal4 driver lines. These dual-purpose lines provide genetic access to specific cell types based on their expression of a native gene while simultaneously mutating one allele of the gene to enable loss-of-function analysis in homozygous animals. While this dual use is often an advantage, the truncation mutations produced by Trojan exons are sometimes deleterious in heterozygotes, perhaps by creating translation products with dominant negative effects. Such mutagenic effects can cause developmental lethality as has been observed with genes encoding essential transcription factors. Given the importance of transcription factors in specifying cell type, alternative techniques for generating specific Gal4 lines that target them are required. Here, we introduce a modified Trojan exon method that retains the targeting fidelity and plug-and-play modularity of the original method but mitigates its mutagenic effects by exploiting the self-splicing capabilities of split inteins. "Split Intein Trojan exons" (siTrojans) ensure that the two truncation products generated from the interrupted allele of the native gene are trans-spliced to create a full-length native protein. We demonstrate the efficacy of siTrojans by generating a comprehensive toolkit of Gal4 and Split Gal4 lines for the segmentally expressed Hox transcription factors and illustrate their use in neural circuit mapping by targeting neurons according to their position along the anterior-posterior axis. Both the method and the Hox gene-specific toolkit introduced here should be broadly useful.
Keywords: Drosophila; cell type; development; genetic access; neural circuit-mapping.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
Similar articles
-
split-intein Gal4 provides intersectional genetic labeling that is repressible by Gal80.Proc Natl Acad Sci U S A. 2023 Jun 13;120(24):e2304730120. doi: 10.1073/pnas.2304730120. Epub 2023 Jun 5. Proc Natl Acad Sci U S A. 2023. PMID: 37276389 Free PMC article.
-
Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes.Cell Rep. 2015 Mar 3;10(8):1410-21. doi: 10.1016/j.celrep.2015.01.059. Epub 2015 Feb 26. Cell Rep. 2015. PMID: 25732830 Free PMC article.
-
Cre-assisted fine-mapping of neural circuits using orthogonal split inteins.Elife. 2020 Apr 14;9:e53041. doi: 10.7554/eLife.53041. Elife. 2020. PMID: 32286225 Free PMC article.
-
Split-inteins and their bioapplications.Biotechnol Lett. 2015 Nov;37(11):2121-37. doi: 10.1007/s10529-015-1905-2. Epub 2015 Jul 8. Biotechnol Lett. 2015. PMID: 26153348 Review.
-
The Drosophila Split Gal4 System for Neural Circuit Mapping.Front Neural Circuits. 2020 Nov 9;14:603397. doi: 10.3389/fncir.2020.603397. eCollection 2020. Front Neural Circuits. 2020. PMID: 33240047 Free PMC article. Review.
Cited by
-
Two classes of amine/glutamate multi-transmitter neurons innervate Drosophila internal male reproductive organs.bioRxiv [Preprint]. 2025 Jul 28:2025.07.23.666348. doi: 10.1101/2025.07.23.666348. bioRxiv. 2025. PMID: 40766443 Free PMC article. Preprint.
-
A collection of split-Gal4 drivers targeting conserved signaling ligands in Drosophila.G3 (Bethesda). 2025 Feb 5;15(2):jkae276. doi: 10.1093/g3journal/jkae276. G3 (Bethesda). 2025. PMID: 39569452 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

