Mutant FOXO1 controls an oncogenic network via enhancer accessibility
- PMID: 38604128
- PMCID: PMC11019358
- DOI: 10.1016/j.xgen.2024.100537
Mutant FOXO1 controls an oncogenic network via enhancer accessibility
Abstract
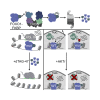
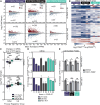
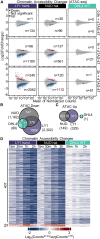
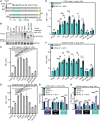
Transcriptional dysregulation is a hallmark of diffuse large B cell lymphoma (DLBCL), as transcriptional regulators are frequently mutated. However, our mechanistic understanding of how normal transcriptional programs are co-opted in DLBCL has been hindered by a lack of methodologies that provide the temporal resolution required to separate direct and indirect effects on transcriptional control. We applied a chemical-genetic approach to engineer the inducible degradation of the transcription factor FOXO1, which is recurrently mutated (mFOXO1) in DLBCL. The combination of rapid degradation of mFOXO1, nascent transcript detection, and assessment of chromatin accessibility allowed us to identify the direct targets of mFOXO1. mFOXO1 was required to maintain accessibility at specific enhancers associated with multiple oncogenes, and mFOXO1 degradation impaired RNA polymerase pause-release at some targets. Wild-type FOXO1 appeared to weakly regulate many of the same targets as mFOXO1 and was able to complement the degradation of mFOXO1 in the context of AKT inhibition.
Keywords: ATAC-seq; DLBCL; FOXO1; PRO-seq; chromatin remodeling; dark zone; enhancer; therapeutics.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.W.H. received research funding from Incyte Inc. through the Vanderbilt-Incyte Alliance during this work. These funds did not support this work.
Figures




Similar articles
-
PAX3-FOXO1 coordinates enhancer architecture, eRNA transcription, and RNA polymerase pause release at select gene targets.Mol Cell. 2022 Dec 1;82(23):4428-4442.e7. doi: 10.1016/j.molcel.2022.10.025. Epub 2022 Nov 16. Mol Cell. 2022. PMID: 36395771 Free PMC article.
-
MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma.Oncotarget. 2015 Jun 20;6(17):15035-49. doi: 10.18632/oncotarget.3729. Oncotarget. 2015. PMID: 25909227 Free PMC article.
-
FOXO1 activation is an effector of SYK and AKT inhibition in tonic BCR signal-dependent diffuse large B-cell lymphomas.Blood. 2016 Feb 11;127(6):739-48. doi: 10.1182/blood-2015-06-654111. Epub 2015 Nov 19. Blood. 2016. PMID: 26585955
-
PAX3-FOXO1: Zooming in on an "undruggable" target.Semin Cancer Biol. 2018 Jun;50:115-123. doi: 10.1016/j.semcancer.2017.11.006. Epub 2017 Nov 14. Semin Cancer Biol. 2018. PMID: 29146205 Review.
-
The regulation of FOXO1 and its role in disease progression.Life Sci. 2018 Jan 15;193:124-131. doi: 10.1016/j.lfs.2017.11.030. Epub 2017 Nov 20. Life Sci. 2018. PMID: 29158051 Review.
Cited by
-
Glucuronolactone Restores the Intestinal Barrier and Redox Balance Partly Through the Nrf2/Akt/FOXO1 Pathway to Alleviate Weaning Stress-Induced Intestinal Dysfunction in Piglets.Antioxidants (Basel). 2025 Mar 18;14(3):352. doi: 10.3390/antiox14030352. Antioxidants (Basel). 2025. PMID: 40227425 Free PMC article.
-
eNRSA: a faster and more powerful approach for nascent transcriptome analysis.Gigascience. 2025 Jan 6;14:giaf071. doi: 10.1093/gigascience/giaf071. Gigascience. 2025. PMID: 40613436 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

