Time-series reconstruction of the molecular architecture of human centriole assembly
- PMID: 38604175
- PMCID: PMC11060037
- DOI: 10.1016/j.cell.2024.03.025
Time-series reconstruction of the molecular architecture of human centriole assembly
Abstract
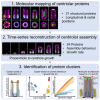
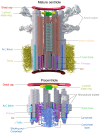
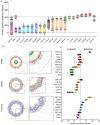
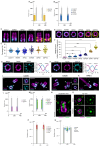
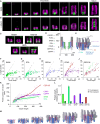
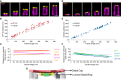
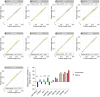
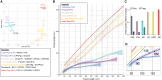
Centriole biogenesis, as in most organelle assemblies, involves the sequential recruitment of sub-structural elements that will support its function. To uncover this process, we correlated the spatial location of 24 centriolar proteins with structural features using expansion microscopy. A time-series reconstruction of protein distributions throughout human procentriole assembly unveiled the molecular architecture of the centriole biogenesis steps. We found that the process initiates with the formation of a naked cartwheel devoid of microtubules. Next, the bloom phase progresses with microtubule blade assembly, concomitantly with radial separation and rapid cartwheel growth. In the subsequent elongation phase, the tubulin backbone grows linearly with the recruitment of the A-C linker, followed by proteins of the inner scaffold (IS). By following six structural modules, we modeled 4D assembly of the human centriole. Collectively, this work provides a framework to investigate the spatial and temporal assembly of large macromolecules.
Keywords: centriole assembly; centriole duplication; centrioles; expansion microscopy; molecular mapping; super-resolution microscopy; time-series reconstruction.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

