Biochemical, structural, and computational analyses of two new clinically identified missense mutations of ALDH7A1
- PMID: 38604394
- PMCID: PMC11073572
- DOI: 10.1016/j.cbi.2024.110993
Biochemical, structural, and computational analyses of two new clinically identified missense mutations of ALDH7A1
Abstract
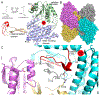
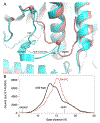
Aldehyde dehydrogenase 7A1 (ALDH7A1) catalyzes a step of lysine catabolism. Certain missense mutations in the ALDH7A1 gene cause pyridoxine dependent epilepsy (PDE), a rare autosomal neurometabolic disorder with recessive inheritance that affects almost 1:65,000 live births and is classically characterized by recurrent seizures from the neonatal period. We report a biochemical, structural, and computational study of two novel ALDH7A1 missense mutations that were identified in a child with rare recurrent seizures from the third month of life. The mutations affect two residues in the oligomer interfaces of ALDH7A1, Arg134 and Arg441 (Arg162 and Arg469 in the HGVS nomenclature). The corresponding enzyme variants R134S and R441C (p.Arg162Ser and p.Arg469Cys in the HGVS nomenclature) were expressed in Escherichia coli and purified. R134S and R441C have 10,000- and 50-fold lower catalytic efficiency than wild-type ALDH7A1, respectively. Sedimentation velocity analytical ultracentrifugation shows that R134S is defective in tetramerization, remaining locked in a dimeric state even in the presence of the tetramer-inducing coenzyme NAD+. Because the tetramer is the active form of ALDH7A1, the defect in oligomerization explains the very low catalytic activity of R134S. In contrast, R441C exhibits wild-type oligomerization behavior, and the 2.0 Å resolution crystal structure of R441C complexed with NAD+ revealed no obvious structural perturbations when compared to the wild-type enzyme structure. Molecular dynamics simulations suggest that the mutation of Arg441 to Cys may increase intersubunit ion pairs and alter the dynamics of the active site gate. Our biochemical, structural, and computational data on two novel clinical variants of ALDH7A1 add to the complexity of the molecular determinants underlying pyridoxine dependent epilepsy.
Keywords: Aldehyde dehydrogenase; Analytical ultracentrifugation; Enzyme kinetics; Missense mutation; Molecular dynamics simulations; X-ray crystallography.
Copyright © 2024 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


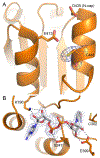



References
-
- Coughlin CR 2nd, Swanson MA 2nd, Spector E Meeks NJL Kronquist KE Aslamy M Wempe MF van Karnebeek CDM Gospe SM Jr. Aziz VG Tsai BP Gao H Nagy PL Hyland K van Dooren SJM Salomons GS Van Hove JLK, The genotypic spectrum of ALDH7A1 mutations resulting in pyridoxine dependent epilepsy: A common epileptic encephalopathy, Journal of inherited metabolic disease, 42 (2019) 353–361. - PMC - PubMed
-
- Tamaura M, Shimbo H, Iai M, Yamashita S, Osaka H, Seizure recurrence following pyridoxine withdrawal in a patient with pyridoxine-dependent epilepsy, Brain Dev, 37 (2015) 442–445. - PubMed
-
- Haidar Z, Jalkh N, Corbani S, Fawaz A, Chouery E, Megarbane A, Atypical pyridoxine dependent epilepsy resulting from a new homozygous missense mutation, in ALDH7A1, Seizure, 57 (2018) 32–33. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

