Multi-Omic Analysis of Esophageal Adenocarcinoma Uncovers Candidate Therapeutic Targets and Cancer-Selective Posttranscriptional Regulation
- PMID: 38604503
- PMCID: PMC11245951
- DOI: 10.1016/j.mcpro.2024.100764
Multi-Omic Analysis of Esophageal Adenocarcinoma Uncovers Candidate Therapeutic Targets and Cancer-Selective Posttranscriptional Regulation
Abstract
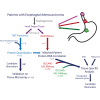
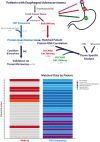
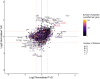
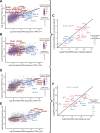
Efforts to address the poor prognosis associated with esophageal adenocarcinoma (EAC) have been hampered by a lack of biomarkers to identify early disease and therapeutic targets. Despite extensive efforts to understand the somatic mutations associated with EAC over the past decade, a gap remains in understanding how the atlas of genomic aberrations in this cancer impacts the proteome and which somatic variants are of importance for the disease phenotype. We performed a quantitative proteomic analysis of 23 EACs and matched adjacent normal esophageal and gastric tissues. We explored the correlation of transcript and protein abundance using tissue-matched RNA-seq and proteomic data from seven patients and further integrated these data with a cohort of EAC RNA-seq data (n = 264 patients), EAC whole-genome sequencing (n = 454 patients), and external published datasets. We quantified protein expression from 5879 genes in EAC and patient-matched normal tissues. Several biomarker candidates with EAC-selective expression were identified, including the transmembrane protein GPA33. We further verified the EAC-enriched expression of GPA33 in an external cohort of 115 patients and confirm this as an attractive diagnostic and therapeutic target. To further extend the insights gained from our proteomic data, an integrated analysis of protein and RNA expression in EAC and normal tissues revealed several genes with poorly correlated protein and RNA abundance, suggesting posttranscriptional regulation of protein expression. These outlier genes, including SLC25A30, TAOK2, and AGMAT, only rarely demonstrated somatic mutation, suggesting post-transcriptional drivers for this EAC-specific phenotype. AGMAT was demonstrated to be overexpressed at the protein level in EAC compared to adjacent normal tissues with an EAC-selective, post-transcriptional mechanism of regulation of protein abundance proposed. Integrated analysis of proteome, transcriptome, and genome in EAC has revealed several genes with tumor-selective, posttranscriptional regulation of protein expression, which may be an exploitable vulnerability.
Keywords: biomarker; esophageal adenocarcinoma; multiomics; proteogenomics; proteomics.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no competing interests.
Figures


References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Edgren G., Adami H.-O., Weiderpass E., Nyrén O., Nyrén O. A global assessment of the oesophageal adenocarcinoma epidemic. Gut. 2013;62:1406–1414. - PubMed
-
- Killcoyne S., Fitzgerald R.C. Evolution and progression of Barrett’s oesophagus to oesophageal cancer. Nat. Rev. Cancer. 2021;21:731–741. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

