Adaptive expansion of ERVK solo-LTRs is associated with Passeriformes speciation events
- PMID: 38605055
- PMCID: PMC11009239
- DOI: 10.1038/s41467-024-47501-3
Adaptive expansion of ERVK solo-LTRs is associated with Passeriformes speciation events
Abstract
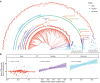
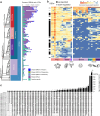
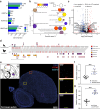
Endogenous retroviruses (ERVs) are ancient retroviral remnants integrated in host genomes, and commonly deleted through unequal homologous recombination, leaving solitary long terminal repeats (solo-LTRs). This study, analysing the genomes of 362 bird species and their reptilian and mammalian outgroups, reveals an unusually higher level of solo-LTRs formation in birds, indicating evolutionary forces might have purged ERVs during evolution. Strikingly in the order Passeriformes, and especially the parvorder Passerida, endogenous retrovirus K (ERVK) solo-LTRs showed bursts of formation and recurrent accumulations coinciding with speciation events over past 22 million years. Moreover, our results indicate that the ongoing expansion of ERVK solo-LTRs in these bird species, marked by high transcriptional activity of ERVK retroviral genes in reproductive organs, caused variation of solo-LTRs between individual zebra finches. We experimentally demonstrated that cis-regulatory activity of recently evolved ERVK solo-LTRs may significantly increase the expression level of ITGA2 in the brain of zebra finches compared to chickens. These findings suggest that ERVK solo-LTRs expansion may introduce novel genomic sequences acting as cis-regulatory elements and contribute to adaptive evolution. Overall, our results underscore that the residual sequences of ancient retroviruses could influence the adaptive diversification of species by regulating host gene expression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Genomic Landscape of Long Terminal Repeat Retrotransposons (LTR-RTs) and Solo LTRs as Shaped by Ectopic Recombination in Chicken and Zebra Finch.J Mol Evol. 2016 Jun;82(6):251-63. doi: 10.1007/s00239-016-9741-0. Epub 2016 May 6. J Mol Evol. 2016. PMID: 27154235
-
Investigation of Polymorphisms Induced by the Solo Long Terminal Repeats (Solo-LTRs) in Porcine Endogenous Retroviruses (ERVs).Viruses. 2024 Nov 20;16(11):1801. doi: 10.3390/v16111801. Viruses. 2024. PMID: 39599915 Free PMC article.
-
Endogenous retroviral solo-LTRs in human genome.Front Genet. 2024 Mar 28;15:1358078. doi: 10.3389/fgene.2024.1358078. eCollection 2024. Front Genet. 2024. PMID: 38606358 Free PMC article. Review.
-
Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors?Retrovirology. 2013 Feb 9;10:16. doi: 10.1186/1742-4690-10-16. Retrovirology. 2013. PMID: 23394165 Free PMC article. Review.
-
Molecular diversity and phenotypic pleiotropy of ancient genomic regulatory loci derived from human endogenous retrovirus type H (HERVH) promoter LTR7 and HERVK promoter LTR5_Hs and their contemporary impacts on pathophysiology of Modern Humans.Mol Genet Genomics. 2022 Nov;297(6):1711-1740. doi: 10.1007/s00438-022-01954-7. Epub 2022 Sep 19. Mol Genet Genomics. 2022. PMID: 36121513 Free PMC article.
Cited by
-
Viral influencers: deciphering the role of endogenous retroviral LTR12 repeats in cellular gene expression.J Virol. 2025 Feb 25;99(2):e0135124. doi: 10.1128/jvi.01351-24. Epub 2025 Jan 31. J Virol. 2025. PMID: 39887236 Free PMC article. Review.
-
Phylogenomics Unveils the Complex Evolution of Retroviruses in Birds.Mol Biol Evol. 2025 Jul 1;42(7):msaf171. doi: 10.1093/molbev/msaf171. Mol Biol Evol. 2025. PMID: 40635662 Free PMC article.
References
-
- Coffin, J. M. Structure and classification of retroviruses. In The Retroviridae 19–49 (Springer US, 1992).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

