Spatiotemporal immune atlas of a clinical-grade gene-edited pig-to-human kidney xenotransplant
- PMID: 38605083
- PMCID: PMC11009229
- DOI: 10.1038/s41467-024-47454-7
Spatiotemporal immune atlas of a clinical-grade gene-edited pig-to-human kidney xenotransplant
Abstract
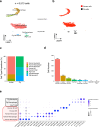
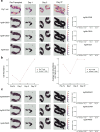
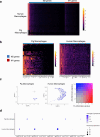
Pig-to-human xenotransplantation is rapidly approaching the clinical arena; however, it is unclear which immunomodulatory regimens will effectively control human immune responses to pig xenografts. Here, we transplant a gene-edited pig kidney into a brain-dead human recipient on pharmacologic immunosuppression and study the human immune response to the xenograft using spatial transcriptomics and single-cell RNA sequencing. Human immune cells are uncommon in the porcine kidney cortex early after xenotransplantation and consist of primarily myeloid cells. Both the porcine resident macrophages and human infiltrating macrophages express genes consistent with an alternatively activated, anti-inflammatory phenotype. No significant infiltration of human B or T cells into the porcine kidney xenograft is detectable. Altogether, these findings provide proof of concept that conventional pharmacologic immunosuppression may be able to restrict infiltration of human immune cells into the xenograft early after compatible pig-to-human kidney xenotransplantation.
© 2024. The Author(s).
Conflict of interest statement
The following authors receive or have received salary support from a research grant from United Therapeutics: R.A., E.D.W., C.F.F., D.E., B.J.O., M.B., G.B., J.P., R.R., S.C.L., A.F.R., J.E.L., and P.M.P. The remaining authors declare no competing interests.
Figures


Update of
-
Spatiotemporal immune atlas of the first clinical-grade, gene-edited pig-to-human kidney xenotransplant.Res Sq [Preprint]. 2023 Jan 9:rs.3.rs-2382345. doi: 10.21203/rs.3.rs-2382345/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Apr 11;15(1):3140. doi: 10.1038/s41467-024-47454-7. PMID: 36711785 Free PMC article. Updated. Preprint.
References
-
- United States Renal Data System. 2020 USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States. (National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, 2020).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

