An RNA-hydrolyzing recombinant minibody prevents both influenza A virus and coronavirus in co-infection models
- PMID: 38605110
- PMCID: PMC11009316
- DOI: 10.1038/s41598-024-52810-0
An RNA-hydrolyzing recombinant minibody prevents both influenza A virus and coronavirus in co-infection models
Abstract
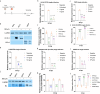
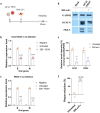
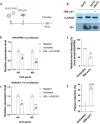
With the lifting of COVID-19 non-pharmaceutical interventions, the resurgence of common viral respiratory infections was recorded in several countries worldwide. It facilitates viral co-infection, further burdens the already over-stretched healthcare systems. Racing to find co-infection-associated efficacy therapeutic agents need to be rapidly established. However, it has encountered numerous challenges that necessitate careful investigation. Here, we introduce a potential recombinant minibody-associated treatment, 3D8 single chain variable fragment (scFv), which has been developed as a broad-spectrum antiviral drug that acts via its nucleic acid catalytic and cell penetration abilities. In this research, we demonstrated that 3D8 scFv exerted antiviral activity simultaneously against both influenza A viruses (IAVs) and coronaviruses in three established co-infection models comprising two types of coronaviruses [beta coronavirus-human coronavirus OC43 (hCoV-OC43) and alpha coronavirus-porcine epidemic diarrhea virus (PEDV)] in Vero E6 cells, two IAVs [A/Puerto Rico/8/1934 H1N1 (H1N1/PR8) and A/X-31 (H3N2/X-31)] in MDCK cells, and a combination of coronavirus and IAV (hCoV-OC43 and adapted-H1N1) in Vero E6 cells by a statistically significant reduction in viral gene expression, proteins level, and approximately around 85%, 65%, and 80% of the progeny of 'hCoV-OC43-PEDV', 'H1N1/PR8-H3N2/X-31', and 'hCoV-OC43-adapted-H1N1', respectively, were decimated in the presence of 3D8 scFv. Taken together, we propose that 3D8 scFv is a promising broad-spectrum drug for treatment against RNA viruses in co-infection.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Broad-Spectrum Antiviral Activity of 3D8, a Nucleic Acid-Hydrolyzing Single-Chain Variable Fragment (scFv), Targeting SARS-CoV-2 and Multiple Coronaviruses In Vitro.Viruses. 2021 Apr 9;13(4):650. doi: 10.3390/v13040650. Viruses. 2021. PMID: 33918914 Free PMC article.
-
A Therapeutically Active Minibody Exhibits an Antiviral Activity in Oseltamivir-Resistant Influenza-Infected Mice via Direct Hydrolysis of Viral RNAs.Viruses. 2022 May 21;14(5):1105. doi: 10.3390/v14051105. Viruses. 2022. PMID: 35632846 Free PMC article.
-
Preventive Activity against Influenza (H1N1) Virus by Intranasally Delivered RNA-Hydrolyzing Antibody in Respiratory Epithelial Cells of Mice.Viruses. 2015 Sep 21;7(9):5133-44. doi: 10.3390/v7092863. Viruses. 2015. PMID: 26402693 Free PMC article.
-
Human Coronavirus OC43 as a Low-Risk Model to Study COVID-19.Viruses. 2023 Feb 20;15(2):578. doi: 10.3390/v15020578. Viruses. 2023. PMID: 36851792 Free PMC article. Review.
-
Host Receptors of Influenza Viruses and Coronaviruses-Molecular Mechanisms of Recognition.Vaccines (Basel). 2020 Oct 6;8(4):587. doi: 10.3390/vaccines8040587. Vaccines (Basel). 2020. PMID: 33036202 Free PMC article. Review.
Cited by
-
Potential Broad-Spectrum Antiviral Agents: A Key Arsenal Against Newly Emerging and Reemerging Respiratory RNA Viruses.Int J Mol Sci. 2025 Feb 10;26(4):1481. doi: 10.3390/ijms26041481. Int J Mol Sci. 2025. PMID: 40003946 Free PMC article. Review.
-
Circulating miRNAs in the Plasma of Post-COVID-19 Patients with Typical Recovery and Those with Long-COVID Symptoms: Regulation of Immune Response-Associated Pathways.Noncoding RNA. 2024 Sep 2;10(5):48. doi: 10.3390/ncrna10050048. Noncoding RNA. 2024. PMID: 39311385 Free PMC article.
References
-
- Pawlowski, C., et al., SARS-CoV-2 and Influenza Co-infection Throughout the COVID-19 Pandemic: An Assessment of Co-infection Rates and Cohort Characterization. medRxiv, 2022: p. 2022.02.02.22270324.
-
- Bao L, et al. Sequential infection with H1N1 and SARS-CoV-2 aggravated COVID-19 pathogenesis in a mammalian model, and co-vaccination as an effective method of prevention of COVID-19 and influenza. Signal Transduct. Target. Therapy. 2021;6(1):200. doi: 10.1038/s41392-021-00618-z. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

