Feasibility of functional precision medicine for guiding treatment of relapsed or refractory pediatric cancers
- PMID: 38605166
- PMCID: PMC11031400
- DOI: 10.1038/s41591-024-02848-4
Feasibility of functional precision medicine for guiding treatment of relapsed or refractory pediatric cancers
Abstract
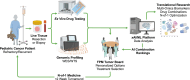
Children with rare, relapsed or refractory cancers often face limited treatment options, and few predictive biomarkers are available that can enable personalized treatment recommendations. The implementation of functional precision medicine (FPM), which combines genomic profiling with drug sensitivity testing (DST) of patient-derived tumor cells, has potential to identify treatment options when standard-of-care is exhausted. The goal of this prospective observational study was to generate FPM data for pediatric patients with relapsed or refractory cancer. The primary objective was to determine the feasibility of returning FPM-based treatment recommendations in real time to the FPM tumor board (FPMTB) within a clinically actionable timeframe (<4 weeks). The secondary objective was to assess clinical outcomes from patients enrolled in the study. Twenty-five patients with relapsed or refractory solid and hematological cancers were enrolled; 21 patients underwent DST and 20 also completed genomic profiling. Median turnaround times for DST and genomics were within 10 days and 27 days, respectively. Treatment recommendations were made for 19 patients (76%), of whom 14 received therapeutic interventions. Six patients received subsequent FPM-guided treatments. Among these patients, five (83%) experienced a greater than 1.3-fold improvement in progression-free survival associated with their FPM-guided therapy relative to their previous therapy, and demonstrated a significant increase in progression-free survival and objective response rate compared to those of eight non-guided patients. The findings from our proof-of-principle study illustrate the potential for FPM to positively impact clinical care for pediatric and adolescent patients with relapsed or refractory cancers and warrant further validation in large prospective studies. ClinicalTrials.gov registration: NCT03860376 .
© 2024. The Author(s).
Conflict of interest statement
N.E.B. is co-founder of and holds shares in First Ascent Biomedical. D.J.A. is co-founder of and holds shares in First Ascent Biomedical. M.F., Z.K., H.A. and O.M.M. are employees of KIDZ Medical Services. The other authors declare no competing interests.
Figures









References
-
- Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin.73, 17–48 (2023). - PubMed
-
- Wong, M. et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat. Med.26, 1742–1753 (2020). - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

